
With increasing global trade there is a requirement for rapid multi-residue screening and quantification methods to efficiently determine residue violations and protect consumers.
In this application note, we describe a single, fast method for confirmation and quantification of more than 425 pesticides. All pesticides were analyzed on a reverse phase column within 12 minutes. One of the objectives of this method was to implement relatively wide MRM windows without any loss in performance. This removes the need to make regular checks on retention time drift, and avoids having to make adjustments to the acquisition method before each analysis.
Two MRM transitions for each pesticide were monitored using both ESI positive and negative modes (deploying polarity switching). The excellent performance of the method has been demonstrated at very low concentrations in chili powder, a very complex matrix. Standard curves in solvent and matrix, easy-to-use instrument and software features to calculate the incurred residues in a chili sample, and data showing robustness after a large number of injections are presented.
Hundreds of compounds are routinely used for crop protection across the globe. With increasing global trade there is a requirement for rapid multi-residue screening and quantification methods to efficiently determine residue violations and protect consumers. Effective multi-residue methods rely on management of the acquisition of a large number of MRM transitions. Setting up overlapping MRM windows based around the retention time of each analyte ensures that no time is wasted acquiring other transitions for compounds that have yet to elute. This optimizes the time spent acquiring data to maximize sensitivity while ensuring a sufficient number of data points across peaks to give good precision. One of the objectives of this method was to implement relatively wide MRM windows without any loss in performance. This removes the need to make regular checks on retention time drift, and avoids having to make adjustments to the acquisition method before each analysis.
In this application note, we describe a single, fast method for confirmation and quantification of more than 425 pesticides. All pesticides were analyzed on a reverse phase column within 12 minutes. Two MRM transitions for each pesticide were monitored using both ESI positive and negative modes (deploying polarity switching). The excellent performance of the method has been demonstrated at very low concentrations in chili powder, which is a very complex matrix. Standard curves in solvent and matrix, easy-to-use instrument and software features to calculate the incurred residues in a chili sample, and data showing robustness after a large number of injections will be presented.
|
LC system: |
ACQUITY UPLC H-Class |
|
Column: |
ACQUITY BEH C18, 1.7 µm, 2.1 x 100 mm |
|
Column temp.: |
45 °C |
|
Injection volume: |
1 µL |
|
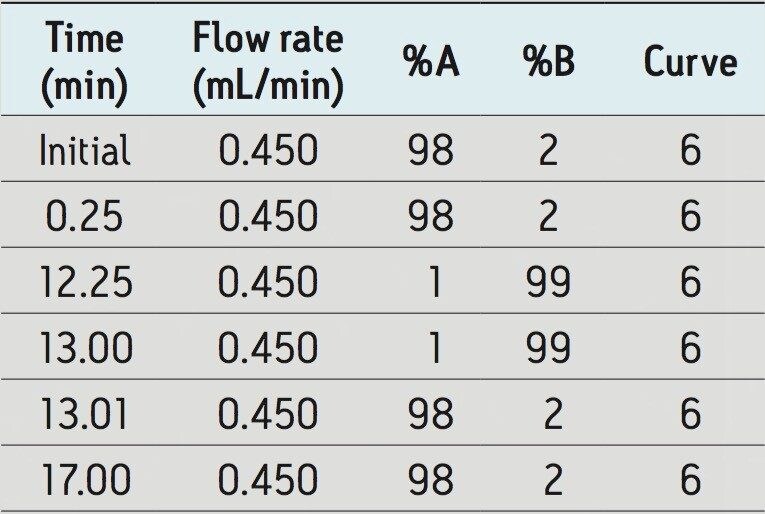
Flow rate: |
0.45 mL/min |
|
Mobile phase A: |
10 mM Ammonium Acetate, pH 5, in water |
|
Mobile phase B: |
10 mM Ammonium acetate in methanol |
|
Weak needle wash: |
50/50 Water/methanol (v/v) |
|
Strong needle wash: |
90/10 Methanol/water (v/v) |
|
Seal wash: |
90/10 Water/methanol |
|
MS system: |
Xevo TQ-S micro |
|
Ionization mode: |
ESI +/- |
|
Capillary voltage: |
1 kV(+) and 0.5 kV(-) |
|
Desolvation temp.: |
500 °C |
|
Desolvation gas flow: |
1000 L/hr |
|
Source temp.: |
150 °C |
|
Acquisition: |
Multiple Reaction Monitoring (MRM) |
Two MRM transitions for each pesticide were generated using Quanpedia, except for fipronil, where 1 MRM transition was used. Methods are shown in Table 2. The data were acquired using MassLynx Software, and processed using TargetLynx XS Application Manager.
Waters LC Multiresidue Pesticide Standards Kit (p/n: 186007574) was used to make a mix of standards. The standards kit has a collection of 204 compounds in 10 different ampoules at individual concentrations of 100 µg/mL. The stock solution of 10 µg/mL was prepared by combining 100 µL from each ampoule. The working standards were further diluted with acetonitrile.
For the solvent calibration curve and standard addition work, acetonitrile was spiked with 204 pesticides at 1, 5, 10, 25, 50, 100, 250, 500, and 1000 µg/kg (ppb) (concentrations equate to sample).
The sample preparation followed the protocol described in a previous application note.1 A DisQuE QuEChERS (CEN method 15662)2 was used to prepare all samples. Briefly, two grams of chili powder was weighed into a centrifuge tube. The sample was mixed with 8 mL of water and vortexed for 30 seconds.
The mixture was extracted with 10 mL of acetonitrile followed by the addition of QuEChERS CEN material (4 g MgSO4, 1 g NaCl and 1.5 g sodium citrate). The resulting mixture was shaken for 1 minute. The tube was then centrifuged at 4000 rpm for 5 minutes, and the supernatant was placed into vials for analysis.
For the matrix match spiked (MMS) calibration, a chili sample was spiked with 204 pesticides at the same level as the solvent calibration range. These spiked levels equaled the concentrations in the chili sample.
The chili sample extract was analyzed for the presence of pesticides using an ACQUITY UPLC H-Class System coupled with the Xevo TQ-S micro Mass Spectrometer. Novel SpaceWire technology facilitates faster acquisition speeds with Xcellerate Ion Transfer (XIT). To achieve the additional sensitivity, the instrument is integrated with StepWave Ion Guide Technology. StepWave effectively removes the neutral molecules, providing additional sensitivity, and improving robustness. Figure 1 shows the Xevo TQ-S micro, along with the StepWave ion guide.
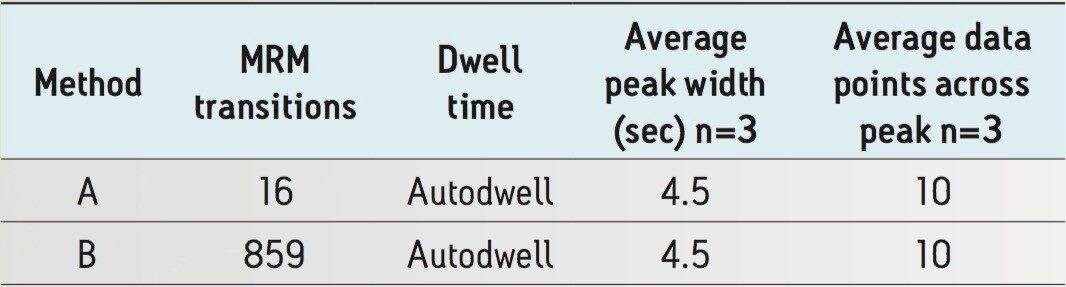
In order to demonstrate the fast acquisition rate and excellent data quality of the MS instrument, a chili extract was post spiked with pesticides at 10 µg/kg (concentration equates to sample) and analyzed using Methods A and B, as described in Table 2. Method A contains 8 pesticides having two MRM transitions for each compound (16 MRMs total). Method B contains 430 pesticides (1 SRM, and 429 with two MRM transitions, totaling 859 MRMs). Both methods were enabled with AutoDwell functionality, at the click of a button. AutoDwell allows MassLynx MS Software to optimize the dwell time automatically for each compound depending on its retention time, as well as the peak width and required data points across the peak, as defined by the user. In this experiment, 1-minute wide acquisition windows were selected for both methods which eliminates the regular checks for retention time drift (due to matrix interferences) and simplifies inter- and/or intra- laboratory method transfer.
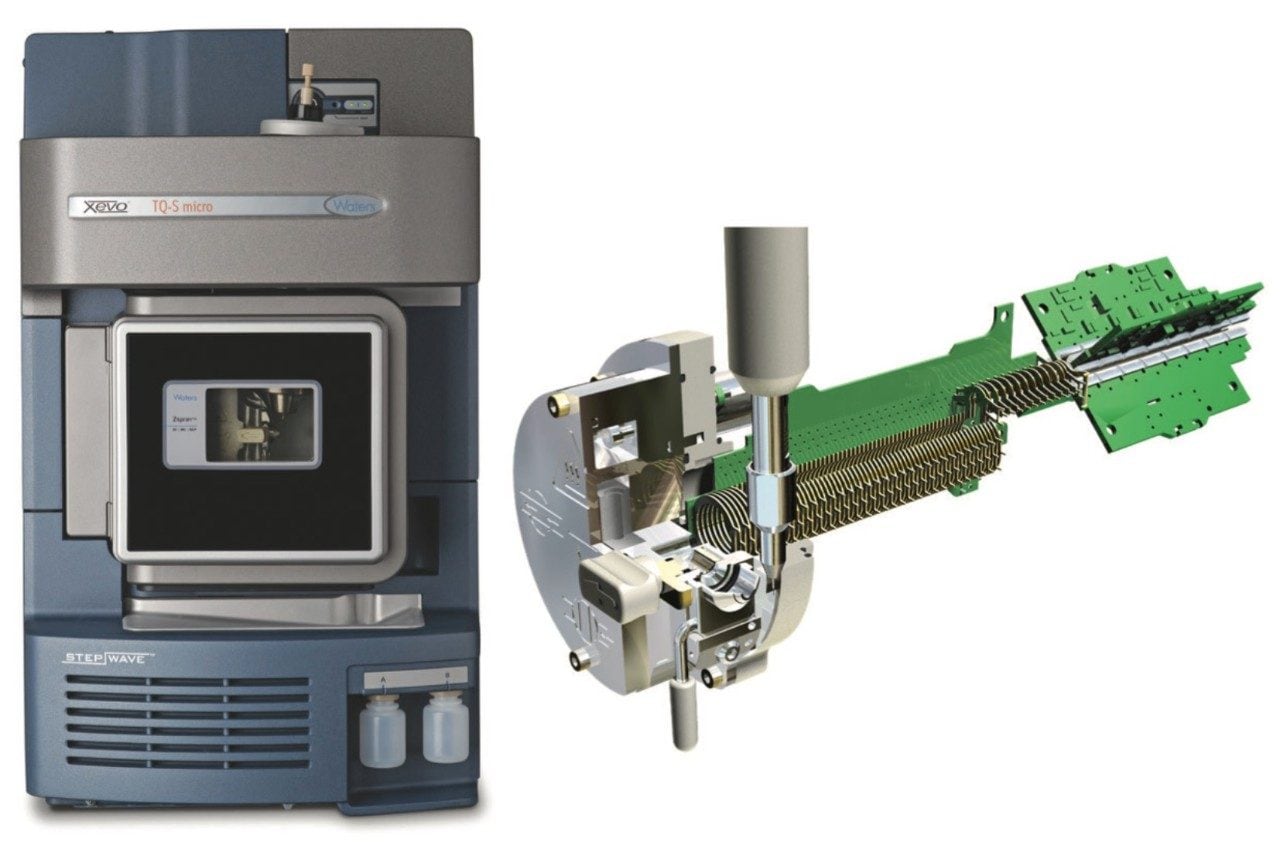
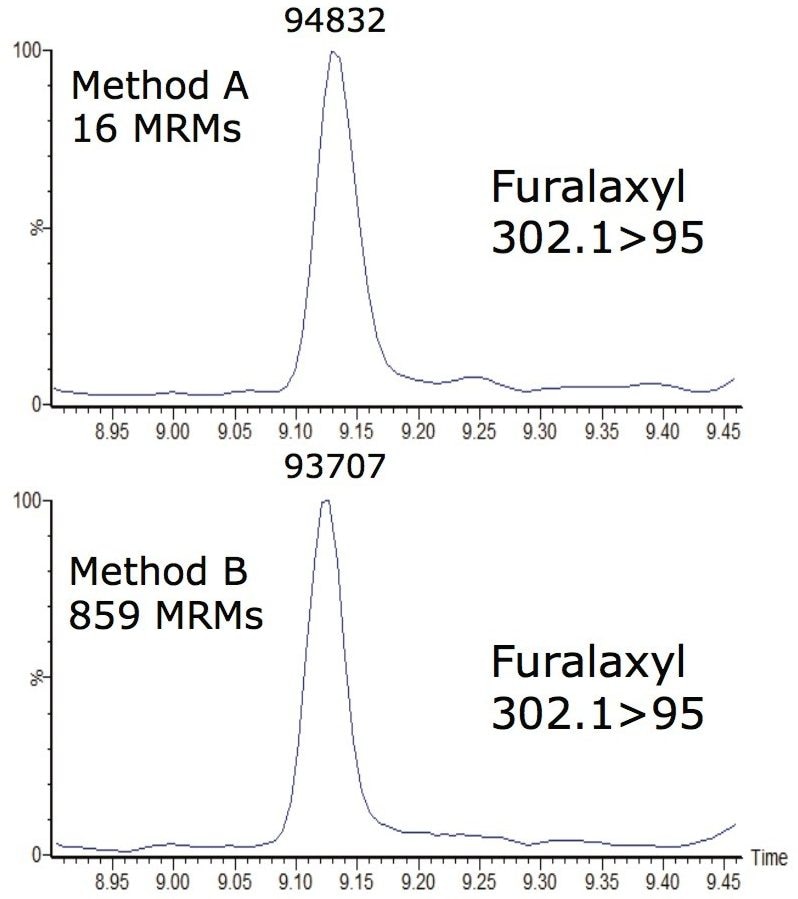
Figure 2 shows a screen shot of part of Method B. In this method, more than 100 pesticides (of the 430 being monitored) eluted between 9 and 10 minutes. Furalaxyl (RT 9.1 min) has eluted within this crowded region. As shown in Table 2, furalaxyl had an average peak width and data points across the peak of 4.4 sec and 10 data points respectively (Method B), for three replicates of the chili sample spiked at 10 µg/kg. Similar results were observed with Method A where fewer transitions (16) were monitored in the method. Despite the large number of compounds in Method B, the data quality was not compromised in the complex matrix.
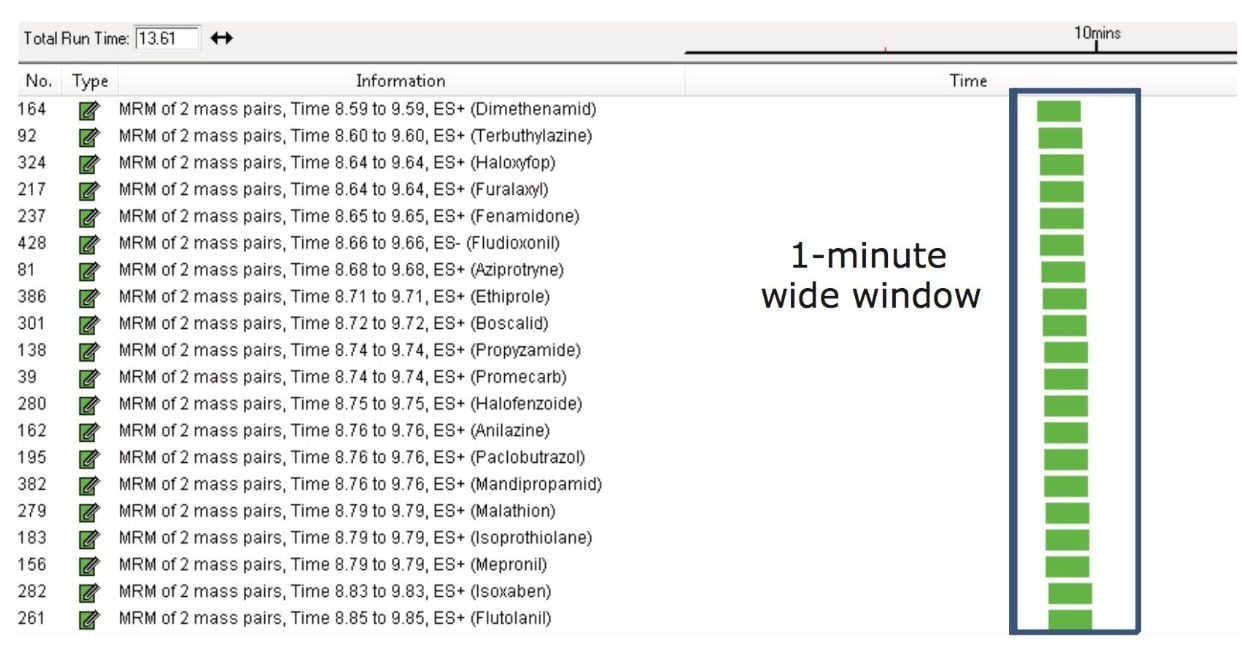
Figure 3 shows chromatograms of furalaxyl acquired by Methods A and B. The peak area difference between Methods A and B which contained 16 and 859 MRM transitions respectively were minimal. Despite Method B having significantly more MRMs than Method A, both methods yielded similar area counts (<2% deviation), which is an acceptable deviation for injection of replicate matrix. Both methods have shown a minimum of 10 data points across the peak, a typical requisite for accurate quantification.
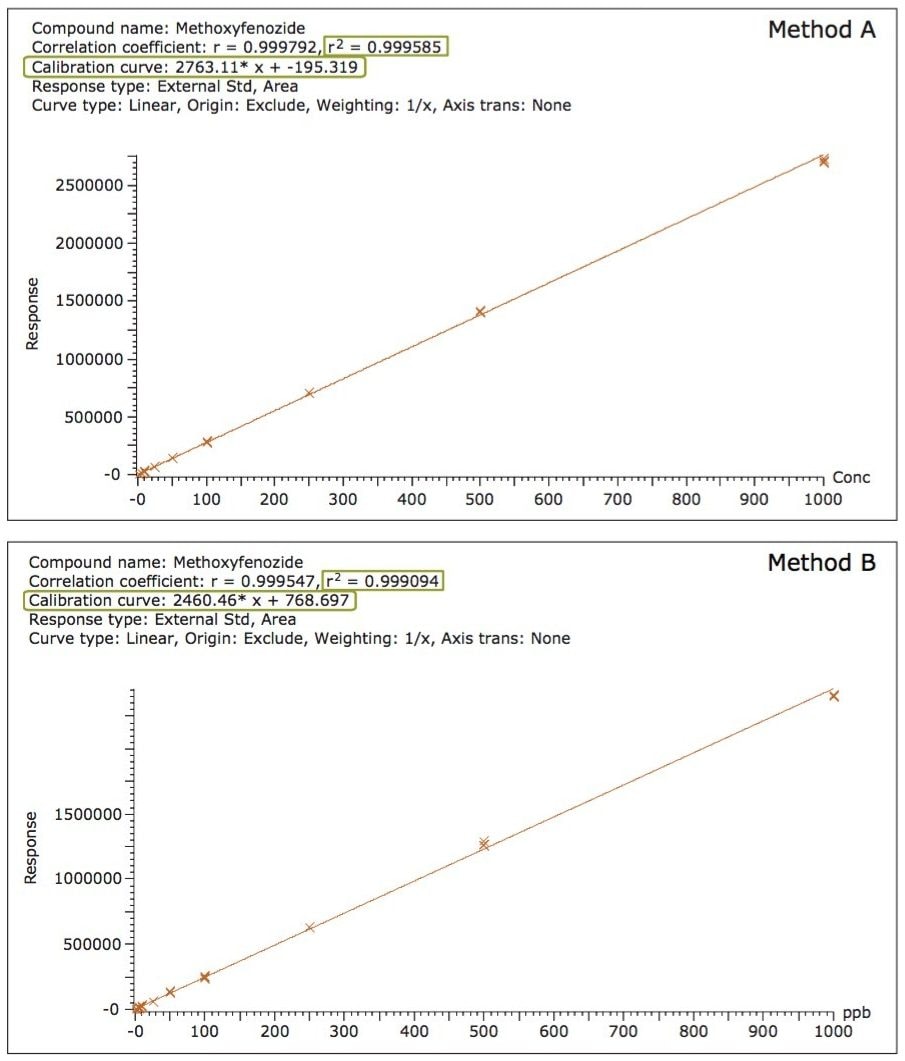
Linearity over the required working range was determined in solvent (data not shown) and matrix matched calibration curves using both MS methods. Figure 4 shows excellent agreement and linearity for matrix matched calibration curve of methoxyfenozide (RT 9.56 min) from 1 to 1000 µg/kg (ppb) using methods A and B. As can be seen in the Figure 4, Methods A and B showed good linearity with an r2 value >0.999 over the entire specified range and similar slope of the equation.
To assess the repeatability and robustness of the method, 300 injections of the spiked chili sample (25 µg/kg) were analyzed by Method B. Figure 5 shows the %RSD of the peak area for example pesticide residues. As shown in Figure 5, all positive and negative ionized compound showed good %RSD (3.3 to 13.9) over 300 injections. Despite the large number of MRM transitions employing polarity switching, Method B showed excellent reproducibility and quantification at a lower concentration in a complex matrix like chili.

A few incurred residues were found when the chili sample was screened with Method B. The standard addition method was employed
to calculate the concentration of incurred residues in the chili sample. Matrix matched samples were prepared using the Auto Addition functionality of the UPLC System, automatically enabling the repeatable mixing of multiple aliquots from several vials within a single injection. The extract was diluted directly in the sample loop before injection. In this study, Auto Addition provided an automated calibration curve for the chili sample by spiking blank chili extract with various concentration of pesticides. Figure 6 shows the Auto Addition setup sample list created in MassLynx Software.
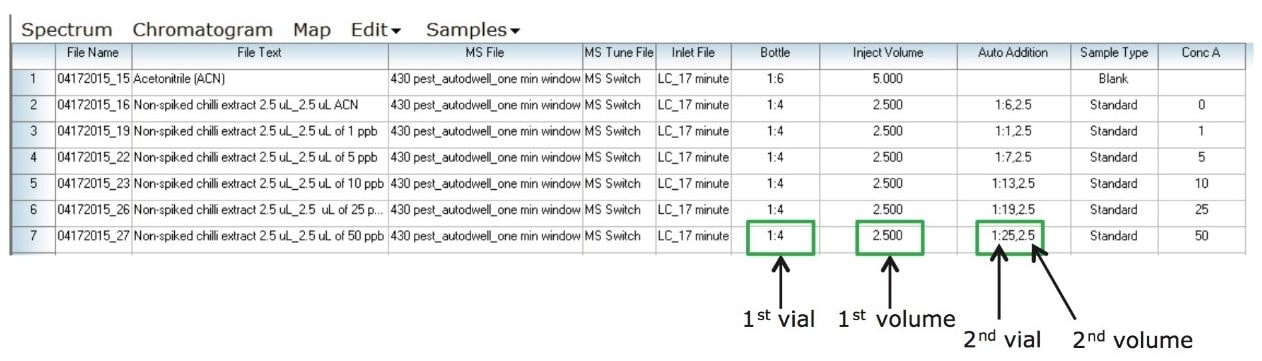
As shown in Figure 6, vials 1:6 and 1:4 contain acetonitrile and a blank chili extract respectively. A mix of pesticide standards in acetonitrile, ranging from 1 to 50 µg/kg (ppb), were placed in individual vials (1:6 to 1:25). Figure 6 also shows the injection order that starts with drawing 2.5 µL of non-spiked extracted chili sample followed by 2.5 µL of acetonitrile. The rest of the injections start by drawing 2.5 µL of the non-spiked extracted chili sample and 2.5 µL of various concentrations of pesticide standards. In this instance, a 5-µL injection total volume was kept constant to ensure accurate and reproducible injections. It also maintains a constant amount of matrix to allow for accurate quantification.
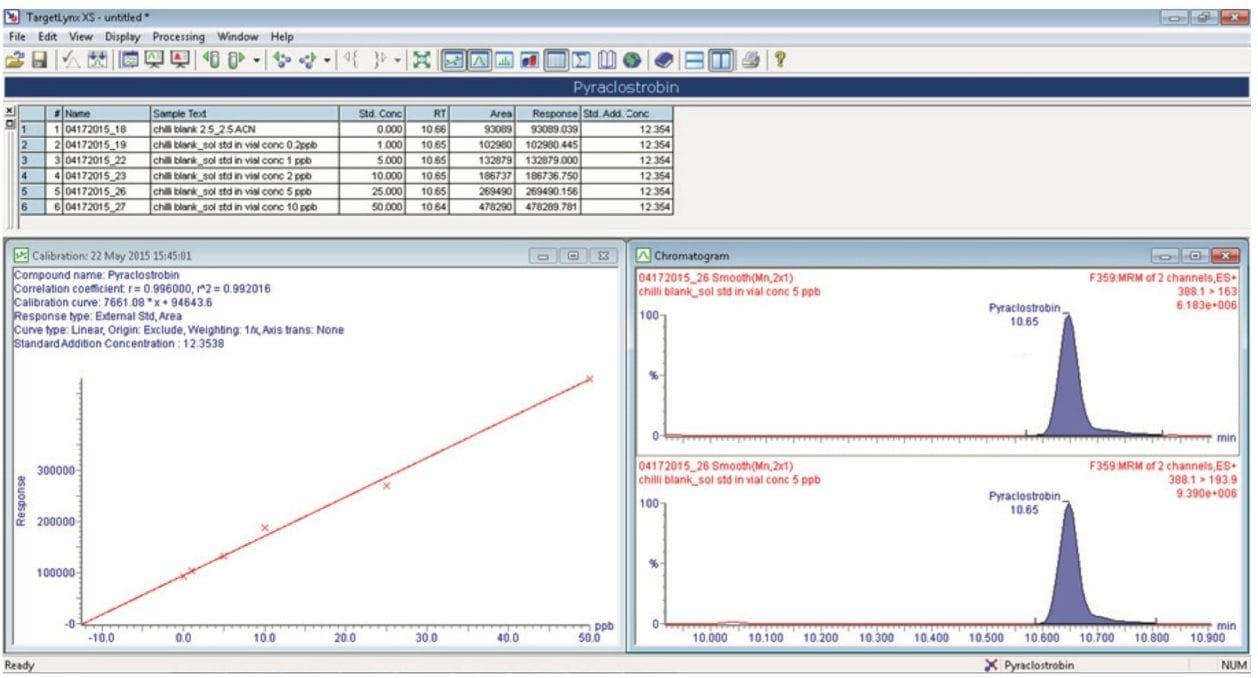
After the data acquisition, incurred residues were then automatically quantified using the Standard Addition functionality in TargetLynx XS Software. Figure 7 shows an example of the calculated concentration of pyraclostrobin (12.35 ppb) in a chili sample using the standard addition approach.
720005559, January 2016