
This is an Application Brief and does not contain a detailed Experimental section.
Characterization of packaging, food contact materials, medical devices, and many other consumables used in various industries is becoming more important due to ever-increasing global regulations.
The initial step in characterizing extractables from packaging includes targeted screening, i.e. testing the extracts for known compounds. This is a well-established process and can be performed in various ways by using analytical techniques ranging from GC-FID-MS to LC-UV-MS. However, the final packaging may have impurities present from starting materials and additional degradants such as those formed during the molding process. The structural elucidation of unknowns is typically a very complex and time-consuming process that requires the analyst to have a high level of expertise.
This technical note describes a simple workflow and methodology for the screening and characterization of cosmetics, 3D printing media, food and pharmaceutical packaging extractable applications.
Characterization of packaging, food contact materials, medical devices, and many other consumables used in various industries is becoming more and more important due to ever-increasing global regulations. The initial step in characterizing extractables from packaging includes targeted screening, i.e. testing the extracts for known compounds. This is a well-established process and can be performed in various ways by using analytical techniques ranging from GC-FID-MS to LC-UV-MS. However, the final packaging may have impurities present from starting materials and additional degradants such as those formed during the molding process. The structural elucidation of unknowns is typically a very complex and time-consuming process that requires the analyst to have a high level of expertise. Waters UNIFI Scientific Information System provides a simple workflow that includes scientific library creation, multivariate statistical analysis, elucidation, and reporting. This single platform Informatics solution enables analysts to evaluate complex data in a more efficient way through simplifying data review and facilitating the decision-making process.
As shown in Figure 1, the workflow starts with a non-targeted, data independent analysis (MSE or HDMSE) acquired on a quadrupole time-of-flight mass spectrometer (QToF) or on an ion mobility QTof mass spectrometer (IMS-QTof). The QTof MS is operated in the alternate scanning MSE mode (where the E represents elevated collision energy), as this technique provides two MS scan functions for data acquisition in one analytical run. The first scan function acquires MS data using low collision energy and collects information on the precursor ions in the sample. For the second scan function the collision energy is ramped from low to high energy which allows for the collection of fragment ions over a wide m/z range. With IMS-QTof, an additional dimension of separation is achieved by the inclusion of ion mobility, thus achieving High Definition Mass SpectrometryE (HDMSE). These types of data acquisitions allow simultaneous collection of precursor and fragment ion information, which is crucial when doing elucidation for unknown compounds. In extractables testing complete information about sample extract is rarely available. Therefore after the targeted screening, the elucidation steps in non-targeted screening are essential.
The sample separation prior to MS analysis can be performed by liquid chromatography using UPLC, by gas chromatography using APGC, as well as by convergence chromatography using UPC2.
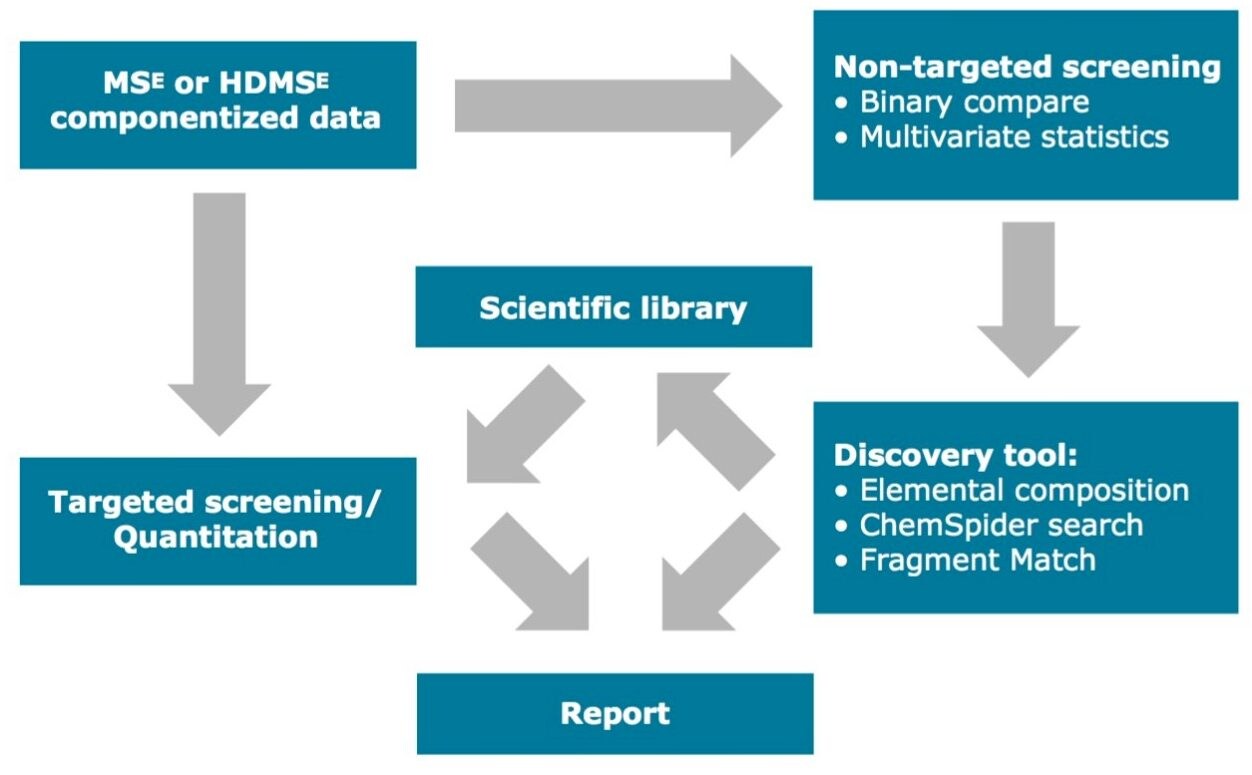
Prior to starting data analysis, the user can create a scientific library based on knowledge of expected compounds in the sample extract, i.e. if the starting compounds in the formulation of a plastic material are known, or a literature search has provided list of compounds that are typically encountered in similar types of packaging. Additionally, regulations provide lists of compounds that are either allowed or prohibited in certain types of packaging, (e.g. food contact materials).
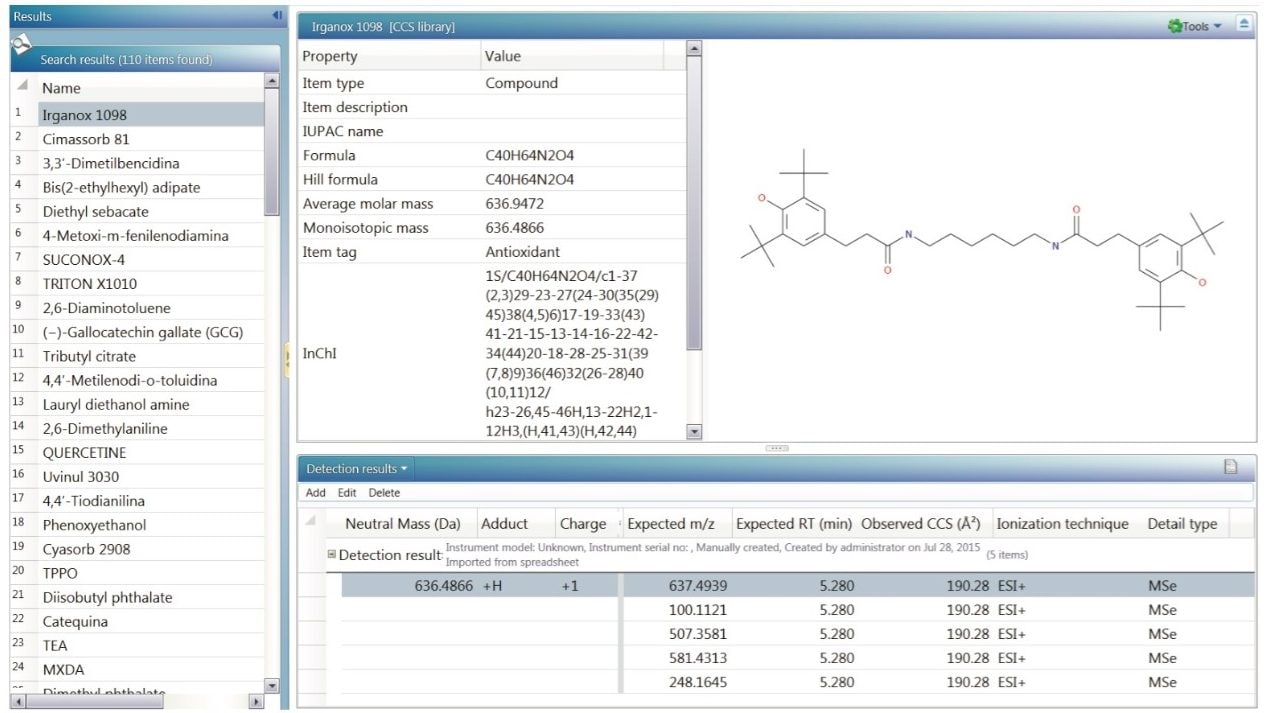
The scientific library (Figure 2.) can contain as much information as is available. The most common information typically included is the compound name, its molecular formula, structure, item tag, and fragmentation information. For ion mobility data, the information needed in screening would be the collisional cross section value (CCS).1 More extensive information about each compound in the library can reduce the number of false positives during targeted screening analysis. Examples of additional information that could be added to a UNIFI scientific library include MS spectra and other relevant documents (e.g. MSDS, articles, SOPs).
Once the data has been acquired, UNIFI uses a target list created by the user from the library to process the raw data and search for compounds which match acceptance criteria. It is also possible to create target lists manually, if required. The user can set up processing criteria such as retention time and mass accuracy tolerances. Subsequently, it is possible to review the proposed identifications based on the number of expected fragments versus the number of expected fragments found, expected and observed CCS values for IMS data with CCS delta (%), isotope intensity matches in ppm or %, among other parameters.
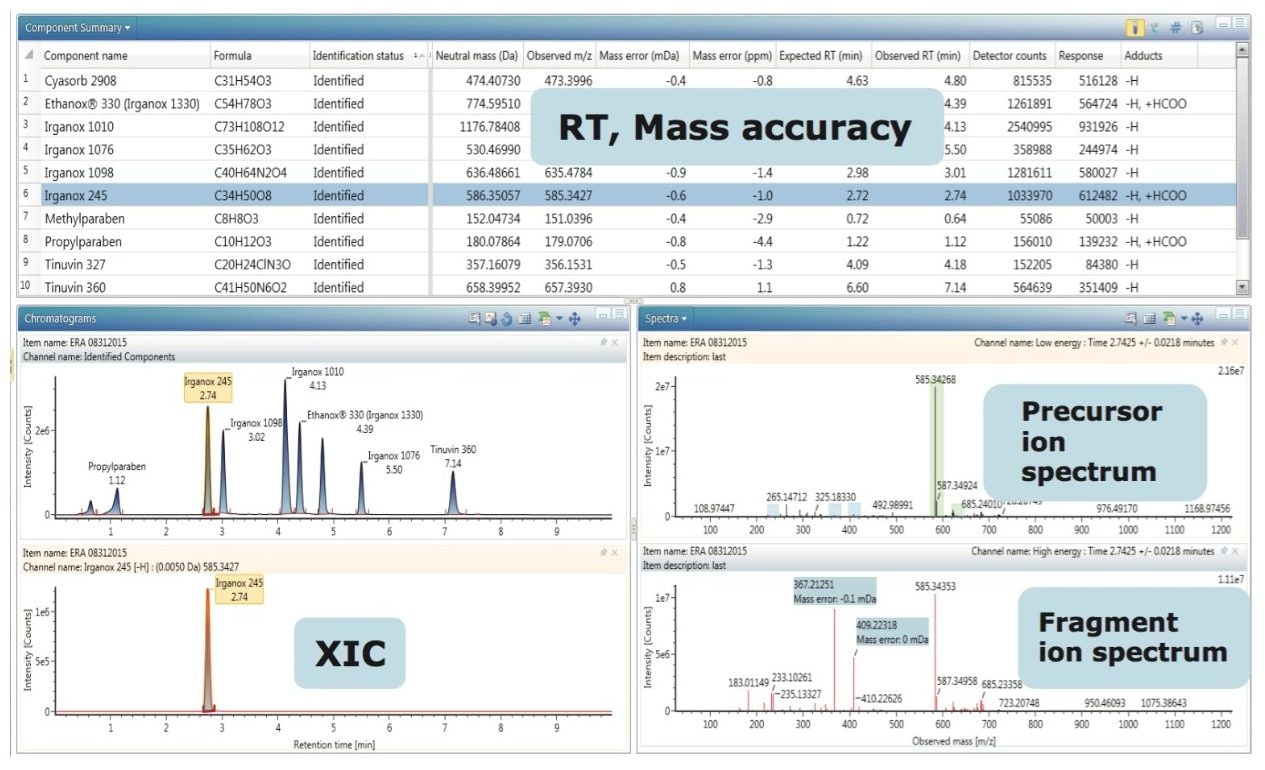
UNIFI allows each user to have a customized workflow which displays information in the preferred dashboard for review (Figure 3). It is possible to review the spectrum for precursor and fragments. Extracted ion chromatograms (XIC) can be displayed for all precursors as well as for fragments. Summary plots can be used to verify the presence or absence, or changes in intensity of a target in other injections. If the appropriate standards have been analyzed during the sample run and a calibration curve is available, it is possible to quantify the identified targets in the analysis.
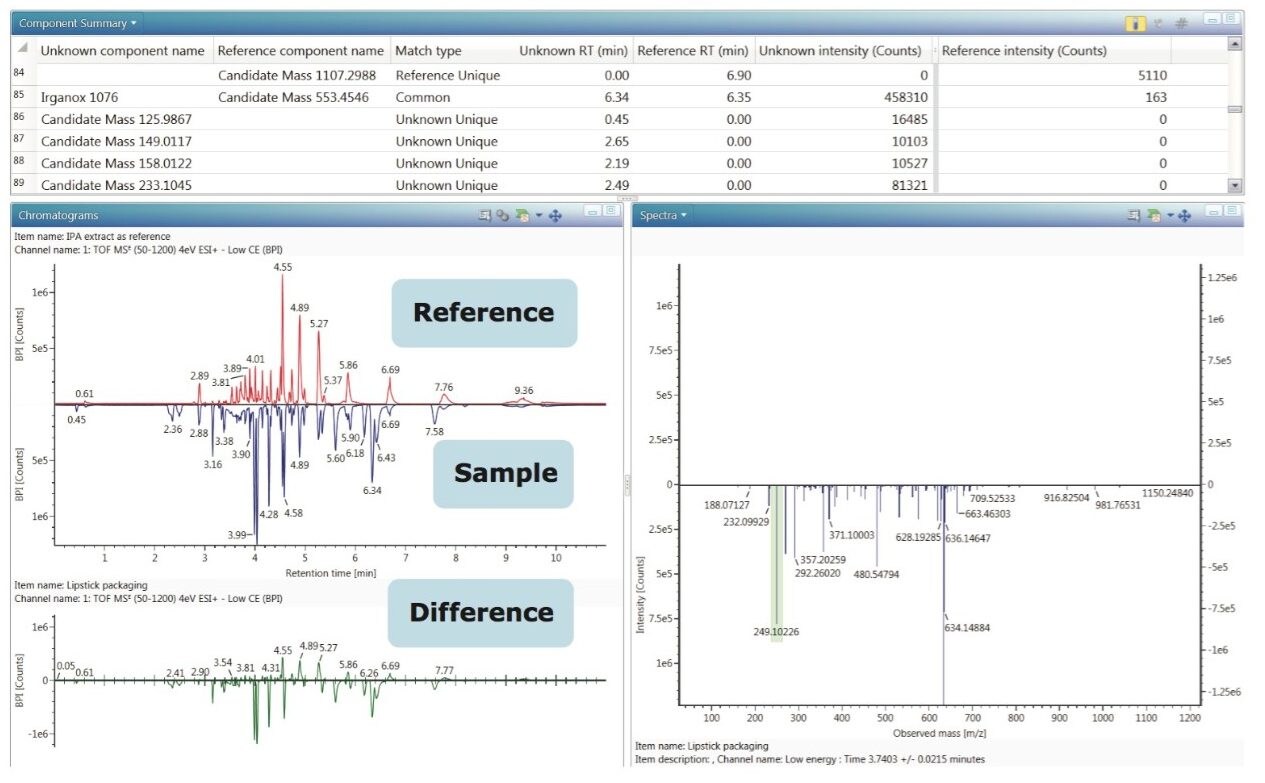
After reviewing the identified targets for false positives and removing them from the identified list, the next question to be answered is “What else is in my sample?” or “What are the differences between a blank extract and sample extract or between these two samples?”. UNIFI has two tools for comparison and statistical analysis. The first one, Binary Compare, allows the user to compare two injections. One injection must be labeled as a reference sample, in this case, an extraction blank.
Masses in the reference spectra and the unknown spectra are considered to be the same component if they are within the specified mass and retention time tolerances. The comparison can be presented graphically as a mirror image of base peak intensity chromatograms (BPI), total ion count chromatograms (TIC), or as a table of candidate masses (Figure 4). Also the spectra of the compound in the reference sample can be displayed in comparison to the unknown sample. The column labeled “Match Type” shows whether the candidate is present only in the unknown sample or in the reference sample, or both. The corresponding match types would be Unknown Unique, Reference Unique, or Common. Typically, compounds that are unique to the sample and absent in the reference sample would be of most interest in an analysis.
If there is a need to compare more than two samples or groups of samples, UNIFI provides Principal Component Analysis (PCA) and other models for data reduction and evaluation by an integrated workflow with statistical software package- EZInfo. PCA is a statistical tool which allows the reduction of a large set of multivariate data into uncorrelated variables called principal components. The differences among the groups of samples are emphasized by Projection to Latent Structures Discriminant Analysis (PLS-DA) model (Figure 5), where a sample group is specified. PLS-DA models the quantitative relationships between the variables X (predictors) and Y (responses) for all of the sample groups. Subsequently, Orthogonal Projection to Latent Structures Discriminant analysis (OPLS-DA) plot demonstrates the differences between two groups.² The data points (markers) in the loadings plot and S-plot are called Accurate Mass/Retention Time pairs (AMRTs). Individual markers that contribute to the biggest differences between the samples can be selected from either the loadings plot or the S-plot and transferred back to the discovery tool for elucidation. When transferring the selected markers, labels can be added to make the data easier to sort and to keep track of markers for different sample groups. When an individual marker is selected from the marker matrix table, a TrendPlot is displayed, allowing the analyst to quickly evaluate its pres-ence in the other samples or injections (Figure 6).

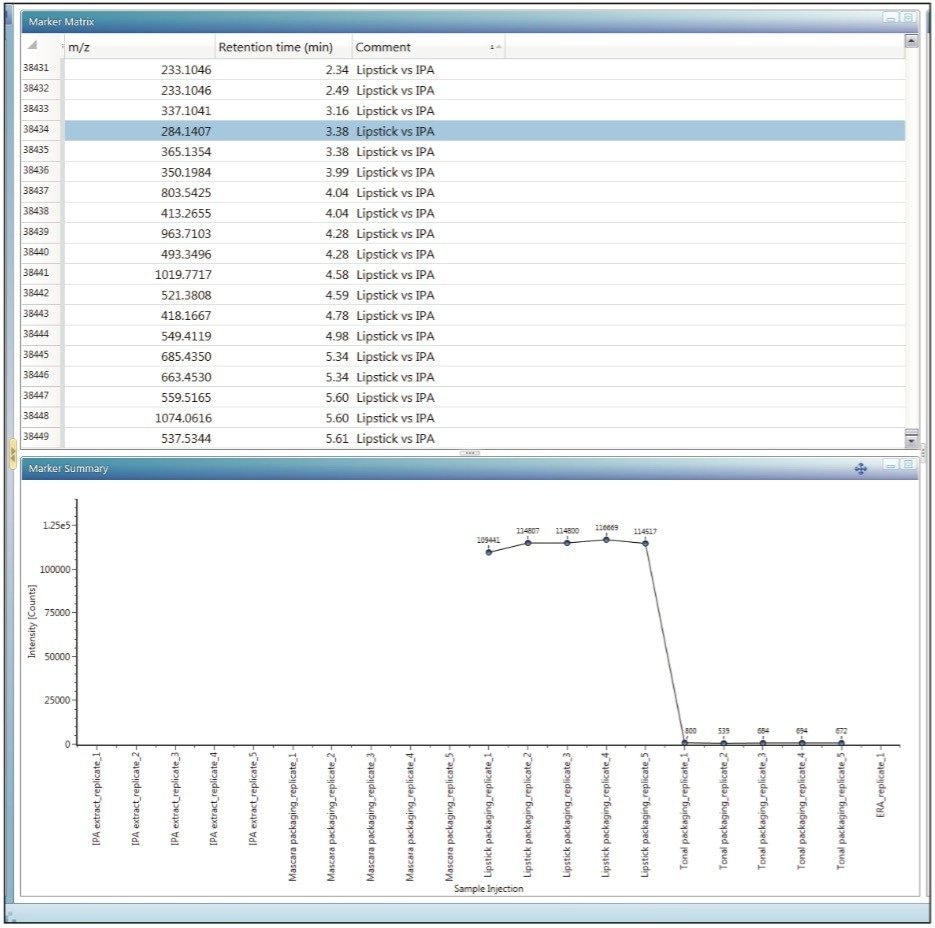
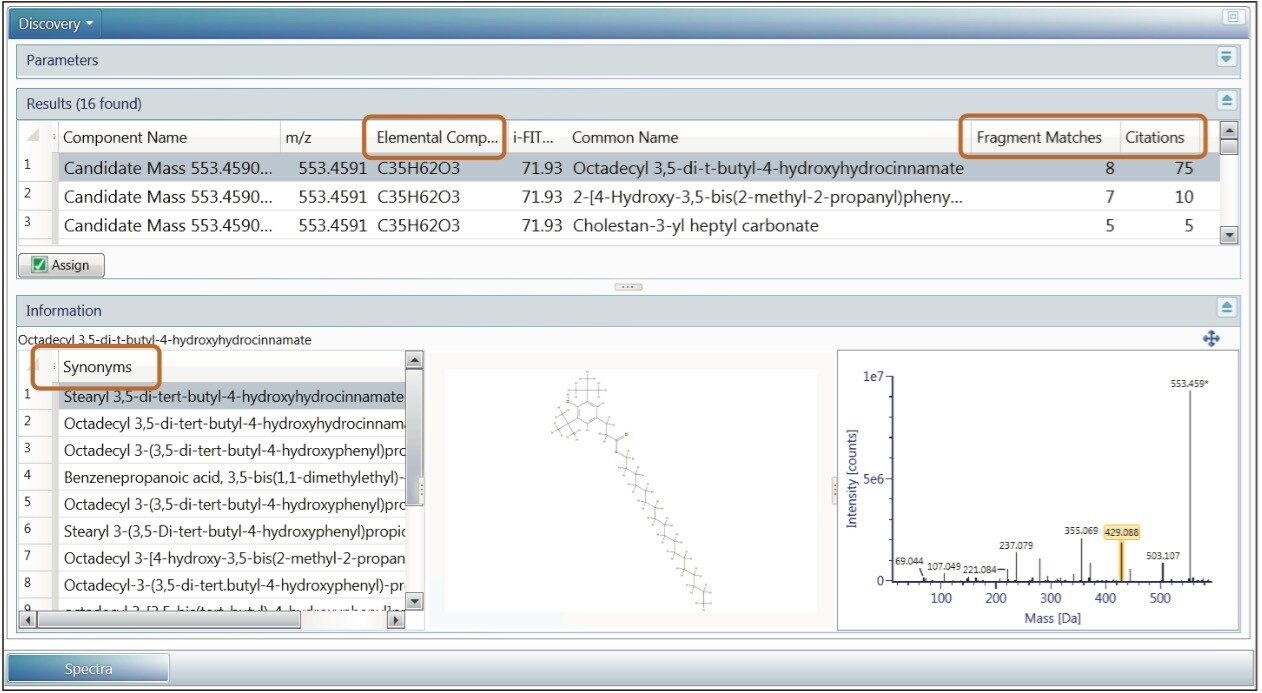
Once the markers are selected either from Binary Compare or from MVA analysis, UNIFI’s Discovery tool can be used to find the possible identity of the ion. The Discovery tool automatically combines all of the analytical information contained in the data: accurate mass, isotope pattern, fragmentation in the high collision energy channel – with the structural database search. The Results table shows the possible molecular formulas for the ion, corresponding structures from a ChemSpider search, and a number of fragments that can be matched to each structure based on fragmentation data. Information returned from the search (Figure 7) also includes the number of citations and synonyms used for each structure. Many polymer additives have common names like Irganoxes and Tinuvins, which helps in further narrowing down the possible compound choices.
When the decision for the compound identity is made, the chosen structure and name can be assigned to the candidate mass ion. Assignment will change the identification status of the candidate to “identified”. All of the elucidated compounds can be added to UNIFI’s scientific library to be used in subsequent targeted screening analysis.
One of the final steps of the analysis is to create a report. A report template can be embedded in the UNIFI Analysis method which can be used for similar types of analysis (Figure 8). The report can be customized to include all the relevant information such as analysis method, processing parameters, chromatograms, spectra, and identified compound summary tables, among others.
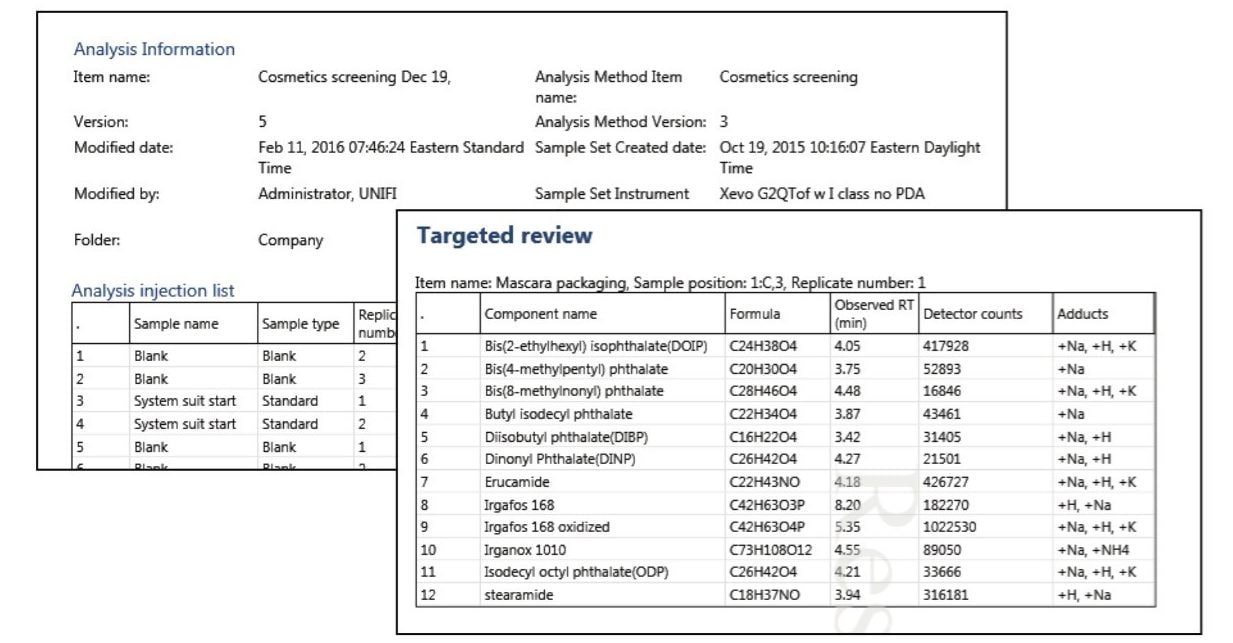
The UNIFI Scientific Information System provides analysts with a well-established workflow for extractable screening analysis. The UNIFI workflow starts with the scientific library for targeted screening, followed by statistical analysis for the determination of markers or relevant compounds. The Discovery tool automatically utilizes information-rich raw data for elemental compositions, followed by a structural database search and fragmentation assignments. This integrated workflow reduces the amount of time required for extractables screening analysis with structural elucidation.
720005688, April 2016