Accurate identification and control of genotoxic impurities, also known as mutagenic impurities, during the development process of a drug substance are critical to product quality and patient safety. Periodic verification testing is an important requirement to demonstrate that the pharmaceutical product meets the acceptable limits and does not pose significant risk to human health.
This application note describes a sensitive and rapid UPLC method coupled with mass detection for the quantitative determination of two genotoxic impurities (GTI) of imatinib mesylate, an anti-neoplastic (anti-cancer) drug used in the treatment of chronic myeloid leukemia and gastrointestinal stromal tumors. Two of its process impurities are classified as potential GTIs and must be monitored during the synthesis process.
The Xevo TQ-S micro method with MRM acquisition mode was developed for the ultra low-level detection of genotoxic impurities of imatinib mesylate API. Utilizing the RADAR function of the MassLynx Software allows acquisition of the full MS scan data simultaneously with the targeted MRM data in a single injection. This enables a quick and accurate assessment of the background matrix for potential presence of additional impurities in the sample or interference with the targeted analytes. The low-detection Xevo TQ-S micro method can be applied for monitoring the fate and purging levels of genotoxic impurities during the development process of the drug substance.
Controlled by a compliant-ready Empower 3 Software, the ACQUITY UPLC H-Class System coupled with an ACQUITY QDa Mass Detector is suitable for routine monitoring of genotoxic impurities in the late stage development of pharmaceutical products through QC environments.
Genotoxic impurities (GTIs) are DNA-reactive substances that may damage DNA, leading to genetic mutations which could potentially cause cancer.1 GTIs, also referred to as mutagenic impurities are unusually toxic. Accurate identification and control of them are critical to ensure product quality and minimize safety risks.
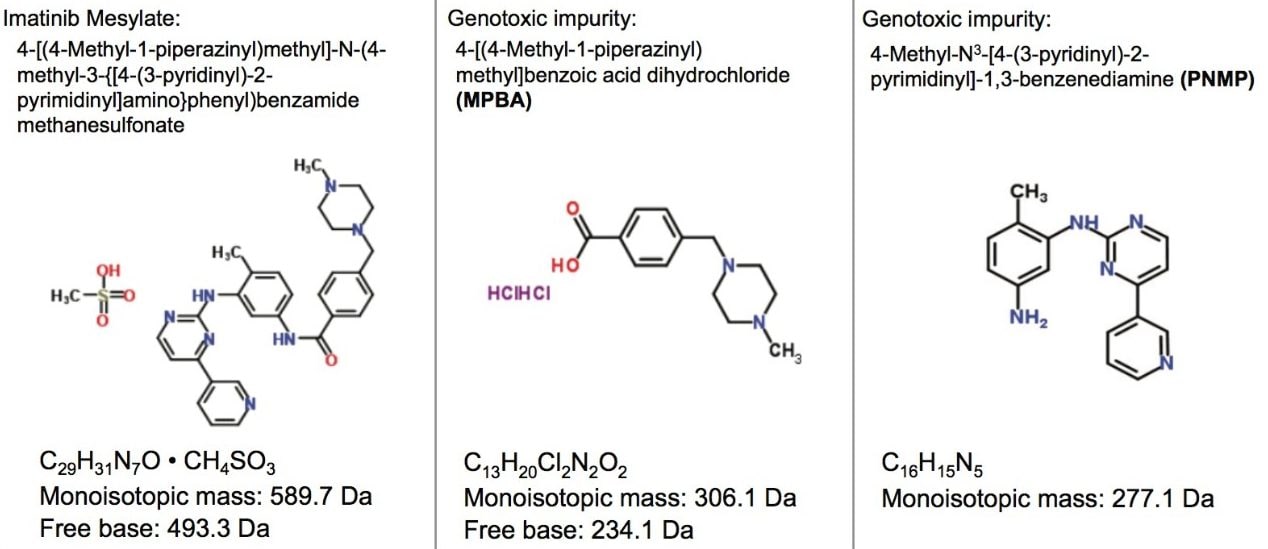
Imatinib mesylate is an anti-neoplastic (anti-cancer) drug used for the treatment of chronic myeloid leukemia and gastrointestinal stromal tumor.2 Two process impurities, 4-[(4-Methyl-1-piperazinyl) methyl]benzoic acid dihydrochloride (MPBA) and 4-Methyl-N3-[4-(3-pyridinyl)-2-pyrimidinyl]-1,3-benzenediamine (PNMP), are classified as potential genotoxic impurities based on the structural alerts (Figure 1), and must be monitored during the synthesis process. The methods reported in literature for quantitative determination of genotoxic impurities in imatinib mesylate utilize HPLC instrumentation and columns, which can be long and consume a high volume of solvents.2,3
Early in API development, there is a need for fast, sensitive, and accurate identification and assessment of impurities to provide guidance on identifying and controlling impurities within acceptable levels. Once impurity profiles are characterized, it becomes equally important to implement methods and technologies that are well suited for accurate, robust, routine, and traceable testing that are critical for late stage development. Typically, early stage development is often performed using information-rich tools (e.g. single/tandem quadrupoles, NMRs), which enable confident characterization of analytes. These methods are then largely implemented as UV-based methodologies in later stages. As drugs become less amenable to UV methods (lower concentrations, more complex formulations), it is becoming increasingly important to establish rapid, MS-friendly workflows to support the entire development life cycle.
The ICH M7 guidance describes several approaches for control of GTIs in drug substances.4 In the case when routing testing is recommended, an analytical method with “as low as reasonably practicable” (ALARP) detection limits will enable accurate monitoring of the fate and purging levels of the impurities during the drug development process. In addition, the ICH M7 recommends periodic verification testing to demonstrate that the product consistently meets specifications.
In this work, we describe a sensitive and rapid UPLC method coupled with mass detection for the quantitative determination of two genotoxic impurities of the imatinib mesylate drug substance. We will investigate the sensitivity and linearity achievable with a tandem quadrupole (Xevo TQ-S micro) mass spectrometer. Then, we will repeat this study on a single quadrupole (ACQUITY QDa) mass detector. We will show that the tandem quadrupole detector is ideal for ultra-sensitive residual detection and quality monitoring of genotoxic impurities in the drug substances, while the ACQUITY QDa provides a robust and sensitive platform suited for routine testing in the late stage development through to QC environments.
The materials used in this study include:
Separate stock solutions of MPBA and PNMP were prepared in methanol at 1.0 mg/mL. An equal volume of each stock solution was transferred to one vial and diluted with standard diluent (20:80 methanol–water) to make a mixture stock solution with 100 µg/mL of each analyte. The mixture solution was serially diluted with standard diluent (20:80 methanol–water) to make linearity standard solutions.
Linearity standards for analysis with Xevo TQ-S micro included the following concentrations: 0.015, 0.1, 1.0, 2.5, 5.0, 7.5, and 10 ng/mL. Linearity standards for detection with an ACQUITY QDa Detector included: 0.25, 1.0, 5.0, 10, 25, 50, 75, and 100 ng/mL.
Imatinib mesylate API samples purchased from three different vendors were prepared in methanol at 5 mg/mL and then diluted with diluent (20:80 methanol–water) to 0.1 mg/mL.
|
LC system: |
ACQUITY UPLC H-Class |
|
Column: |
ACQUITY UPLC HSS PFP, 1.8 µm, 2.1 mm x 50 mm |
|
Column temp.: |
40 °C |
|
Flow Rate: |
0.7 mL/min |
|
Injection Volume: |
8.0 µL |
|
Solvent A: |
2.5 mM ammonium formate in H2O, pH 3.0 adjusted with formic acid |
|
Solvent B: |
Acetonitrile |
|
Purge wash: |
50:50 H2O–CH3OH |
|
Sample wash: |
50:50 H2O–CH3OH |
|
Seal wash: |
90:10 H2O–ACN |
|
Separation: |
Gradient as described in Table 1 |
|
Step |
Time(minutes) |
Solvent A (%) |
Solvent B (%) |
Curve |
|---|---|---|---|---|
|
1.0 |
Initial |
95.0 |
5.0 |
Initial |
|
2.0 |
5.0 |
5.0 |
95.0 |
6.0 |
|
3.0 |
5.5 |
5.0 |
95.0 |
6.0 |
|
4.0 |
5.6 |
95.0 |
5.0 |
6.0 |
|
5.0 |
7.5 |
95.0 |
5.0 |
6.0 |
Table 1. ACQUITY UPLC H-Class mobile phase gradient.
|
Ionization mode: |
ESI+ |
|
Desolvation temp.: |
600 °C |
|
Desolvation gas: |
1000 L/Hr |
|
Capillary voltage: |
0.5 kV |
|
Acquisition: |
MRM with RADAR mode |
The MRM transition ions and ionization parameters for each ion were automatically identified using IntelliStart within the MassLynx Software. IntelliStart automatically tunes the instrument parameters to aid development of MS method for the targeted compounds of interest. The optimized ionization parameters for genotoxic impurities analyzed in this study such as cone voltages and collision gas energies of MRM transition are listed in Table 2.
|
Name |
Transition (m/z) |
Cone Voltage (V) |
Collision Energy (V) |
|
|---|---|---|---|---|
|
MPBA |
235.1 →135.0 |
64.0 |
22.0 |
|
|
PNMP |
278.2 →106.0 |
78.0 |
26.0 |
Table 2. Ionization parameters for MRM transitions.
|
Mass detector: |
ACQUITY QDa, Performance option |
|
Ionization mode: |
ESI+ |
|
MS acquisition time: |
0–5 mins |
|
MS acquisition range: |
100–600 m/z |
|
SIR: |
235.2 and 278.2 Da |
|
Sampling rate: |
10 pts/sec |
|
Capillary voltage: |
0.4 kV (+) |
|
Cone voltage: |
8 V |
|
Probe temp.: |
600 °C |
|
Data: |
Centroid |
The structure and chemical information for genotoxic impurities of imatinib mesylate are shown in Figure 1.
Multiple reaction monitoring (MRM) is a highly sensitive and selective technique for the quantitation of targeted compounds in complex sample matrices. The multiple reaction monitoring (MRM) experiment is performed by defining the parent mass of the targeted compound for MS/MS fragmentation and then monitoring a fragment ion. The MRM experiment is known as a transition and is expressed as parent mass > fragment mass.5
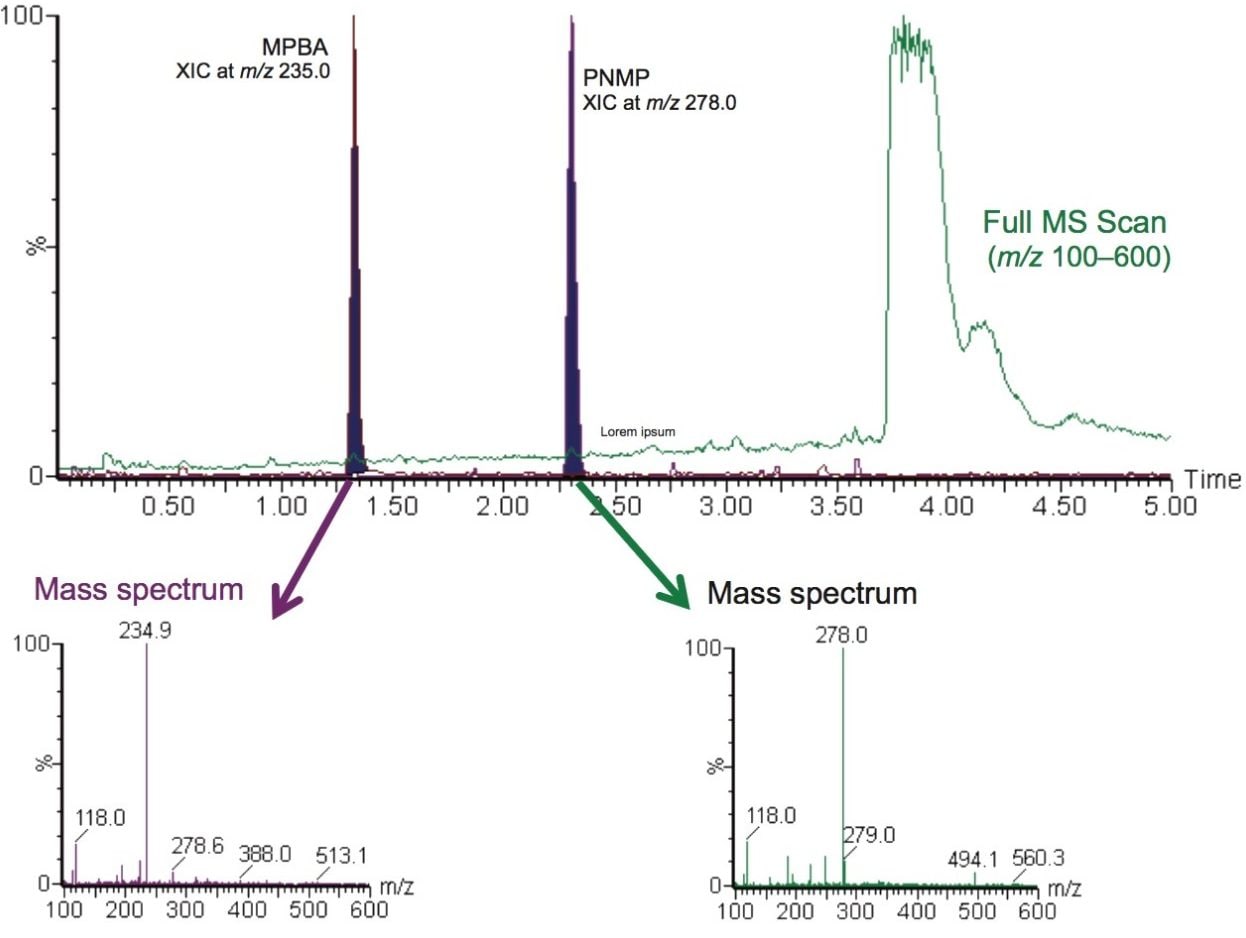
The genotoxic impurities of imatinib mesylate were measured using the MRM acquisition mode and the optimized ionization parameters developed by IntelliStart of the MassLynx Software are described in the experimental section. An example of chromatographic data of MPBA and PNMP acquired with MRM is shown in Figure 2.
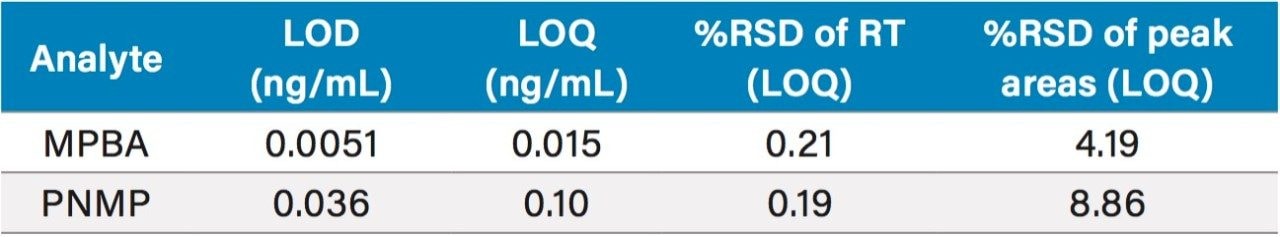
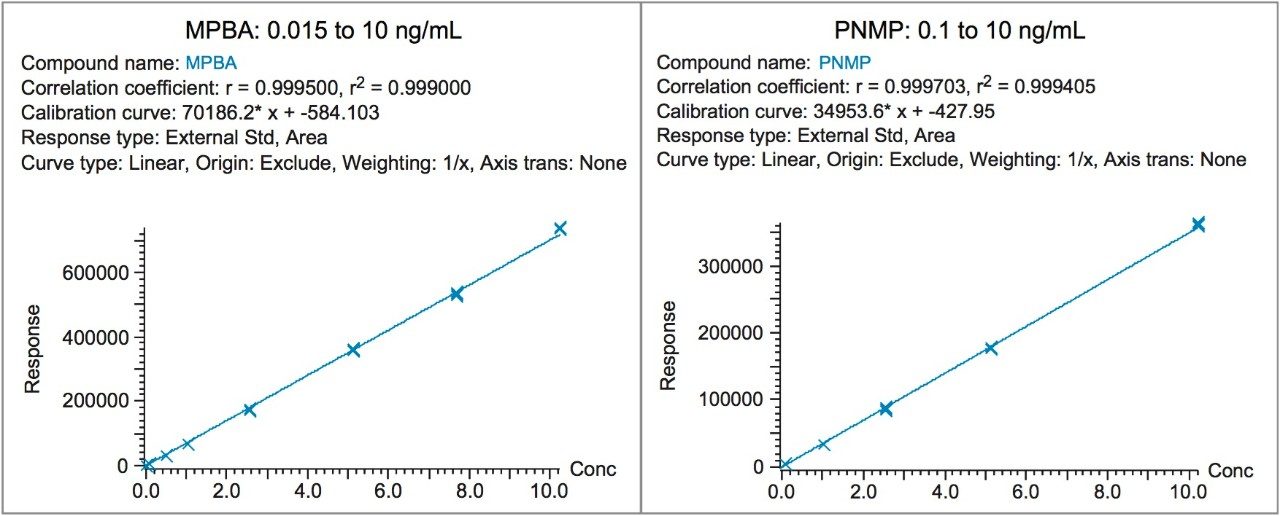
Limit of detection (LOD) and quantification (LOQ) were determined following the signal-to-noise criteria of 3:1 and 10:1, respectively. Data from six replicate injections of standard solutions containing genotoxic impurities was evaluated to demonstrate performance at the LOD and LOQ levels. Results are summarized in Table 3. The LOQ for MPBA and PNMP was found to be 0.015 ng/mL and 0.1 ng/mL, respectively. The %RSD of the peak areas for six replicate injections of the LOQ solution was 8.86%. Linearity of the MS method was evaluated from LOQ to 10 ng/mL range. Both genotoxic impurities exhibited an acceptable linear response with the correlation coefficients of ≥0.999 (Figure 3).

PI samples purchased from three different suppliers were analyzed for presence of the genotoxic impurities, MPBA and PNMP, using MRM mode with RADAR enabled. The RADAR functionally of the Xevo TQ-S micro allows a simultaneous recording of the MS scan acquisition mode over a defined m/z range and multiple MRM transitions. Example of the API 1 sample in the MS full scan collected over the m/z 100–600 range is displayed in Figure 4. The MS scan data enables accurate identification of the sample components and assessment of the background matrix interference by evaluating mass spectrum of the extracted ion chromatogram (XIC) for each analyte. The MS spectrum data shows that the genotoxic impurities are not coeluting with any major background ions.
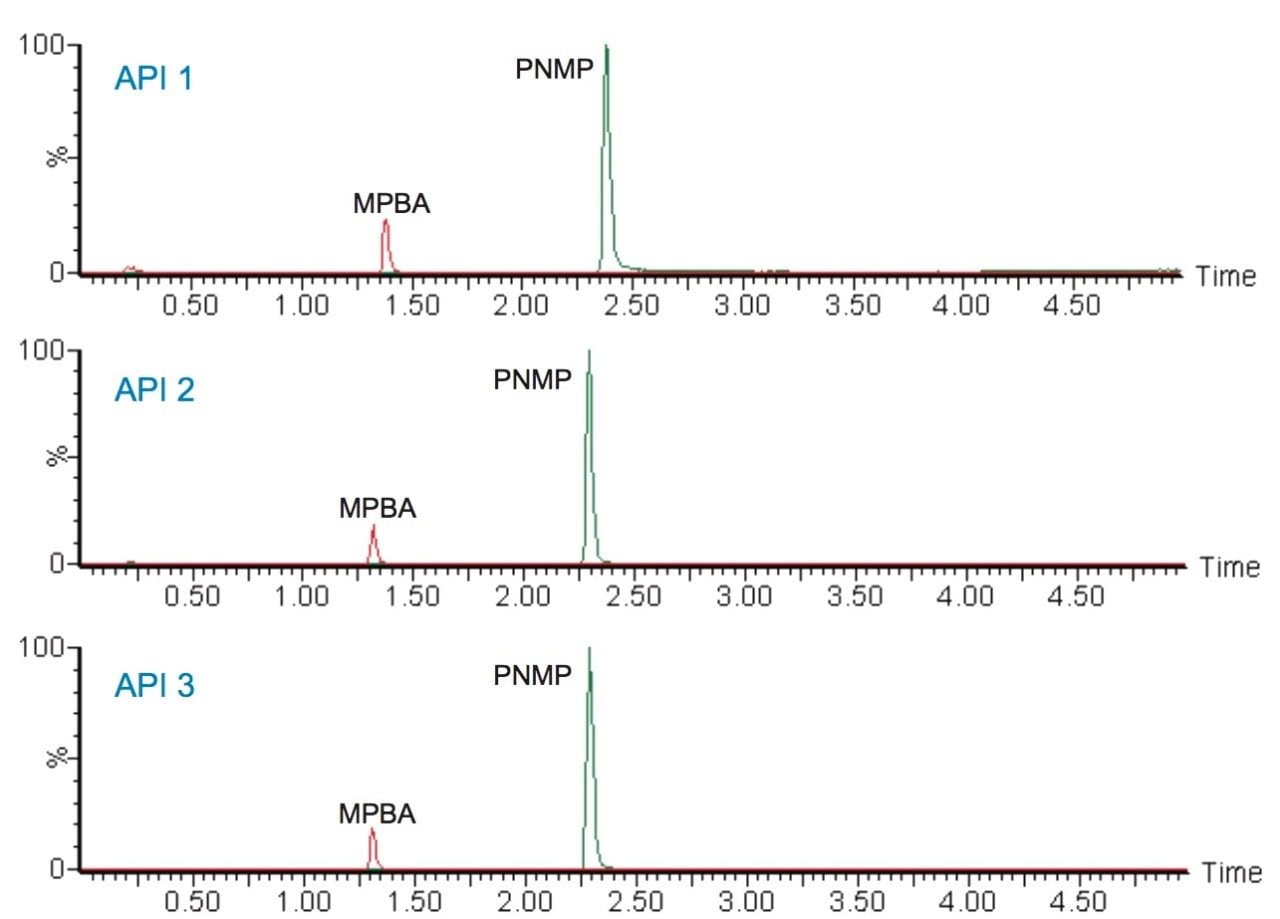
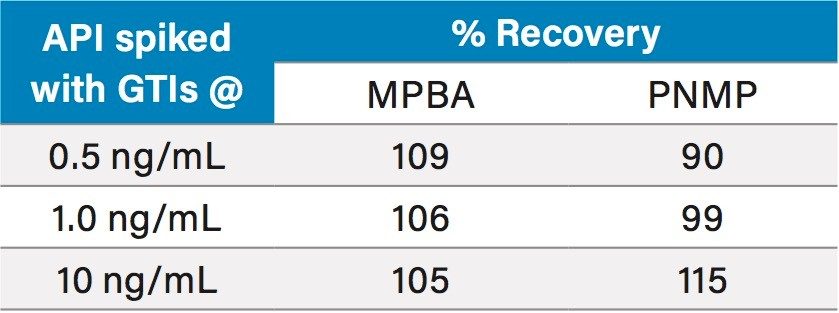
For quantitative analysis of the genotoxic impurities, all three batches of API samples prepared at 0.1 mg/mL were analyzed using MRM mode. The data shows that all three APIs were found to contain both genotoxic impurities (Figure 5). The API 1 sample was spiked with 0.5, 1.0, and 10 ng/mL of genotoxic impurities (n=3 per each level) and analyzed for recovery. The data was quantified against the calibration curve to calculate the amount of MPBA and PNMP. The results generated for the recovery study can be found in Table 4, and ranged from 90 to 115%.
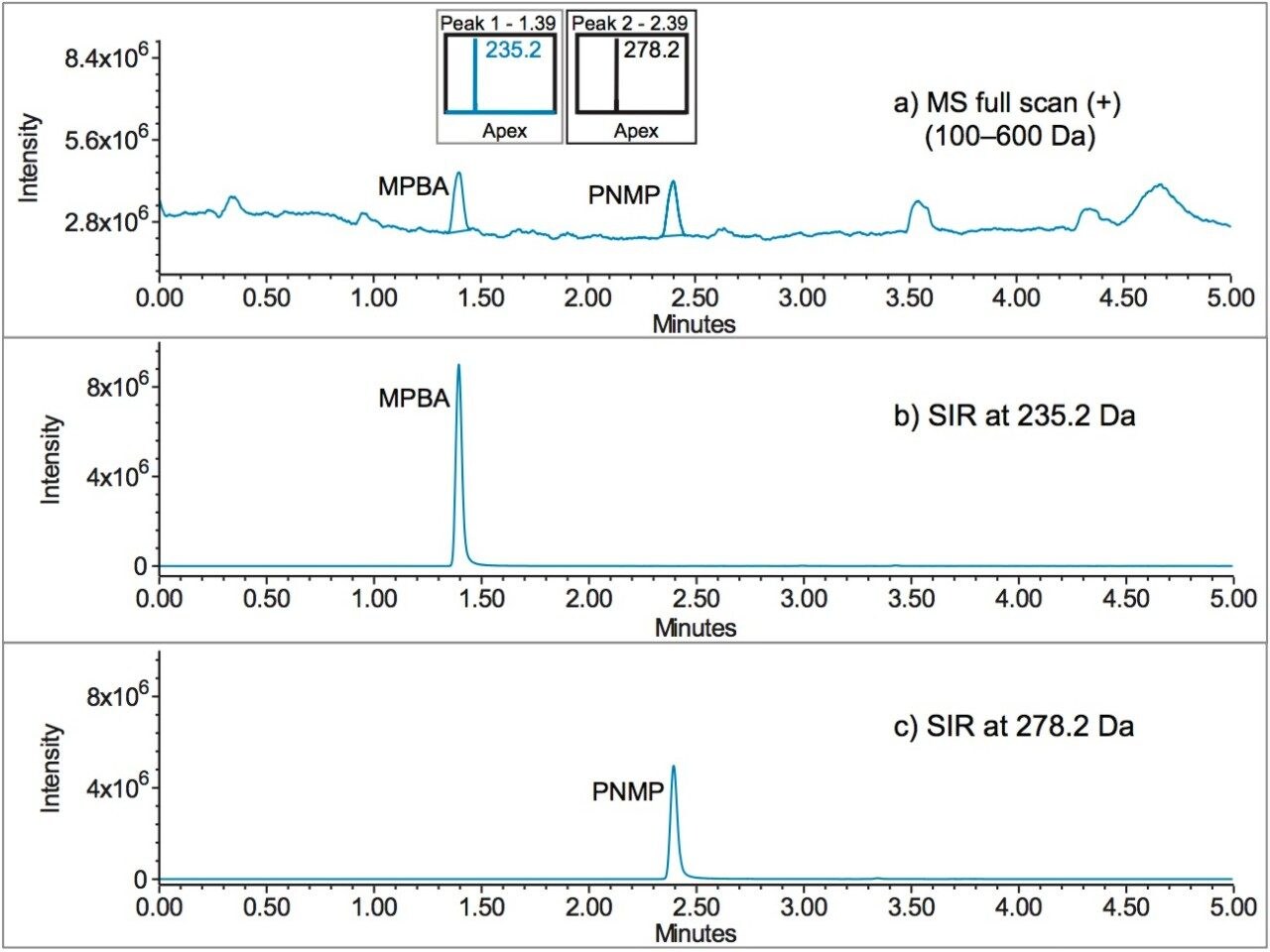
The data acquired using an ACQUITY QDa Mass Detector is displayed in Figure 6. The MS full scan data (Figure 6A) shows all the masses detected across the collected mass range (100–600 Da). For quantitative analysis, the genotoxic impurities (MPBA and PNMP) were measured using a single ion recording (SIR) mode, which only records intensity for a single ion of interest (Figure 6B and 6C). The SIR mode improves method sensitivity and simplifies analysis for targeted compounds.
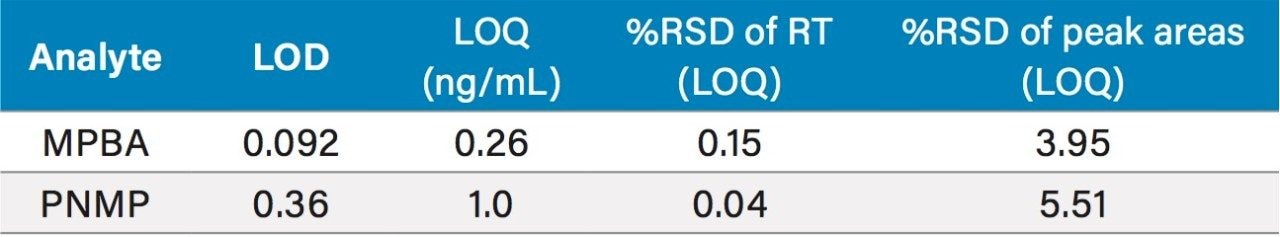
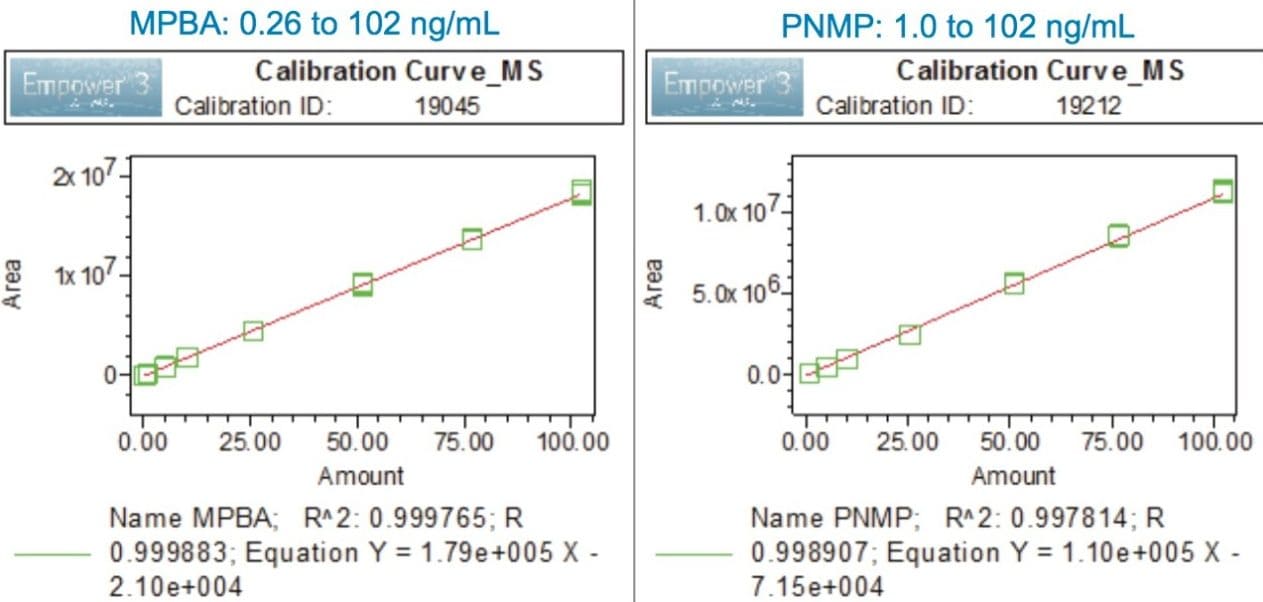
The LOD and LOQ achievable with the ACQUITY QDa were determined using the signal-to-noise data generated from six replicate injections of a standard containing genotoxic impurities (Table 5). The LOQ for MPBA and PNMP was found to be 0.26 ng/mL and 1.0 ng/mL, respectively. The %RSD of the peak areas was less than 5.51%. The method exhibited an acceptable linearity from LOQ to 100 ng/mL range with correlation coefficients of ≥0.997 (Figure 7).
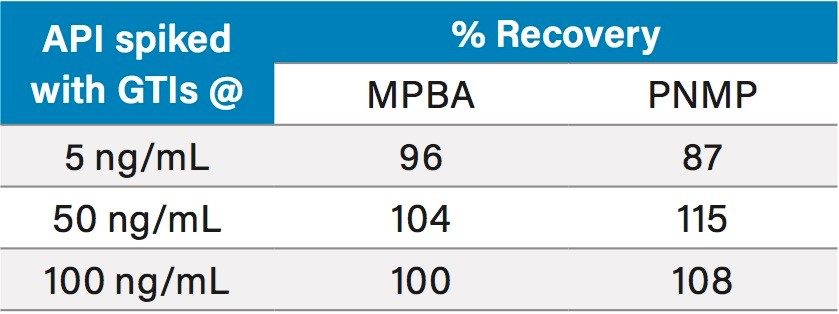
To investigate recovery, the API 1 sample prepared at 0.1 mg/mL was spiked with 5, 50, and 100 ng/mL of MPBA and PNMP (n=3 per each level). The % Recovery results ranged from 87 to 115% (Table 6).
The Xevo TQ-S micro method with MRM acquisition mode was developed for the ultra low-level detection of genotoxic impurities of imatinib mesylate API. Utilizing the RADAR function of the MassLynx Software allows acquisition of the full MS scan data simultaneously with the targeted MRM data in a single injection. This enables a quick and accurate assessment of the background matrix for potential presence of additional impurities in the sample or interference with the targeted analytes. The low-detection Xevo TQ-S micro method can be applied for monitoring the fate and purging levels of genotoxic impurities during the development process of the drug substance.
Controlled by a compliant-ready Empower 3 Software, the ACQUITY UPLC H-Class System coupled with an ACQUITY QDa Mass Detector is suitable for routine monitoring of genotoxic impurities in the late stage development of pharmaceutical products through QC environments.
Overall, accurate identification and control of genotoxic impurities during the development process of a drug substance are critical to product quality and patient safety. Periodic verification testing is an important requirement to demonstrate that the pharmaceutical product meets the TTC-based acceptable limits and does not pose significant risk to human health.
720005873, January 2017