
Forced degradation is a critical analytical study for the development of stability-indicating methods used by pharmaceutical companies as part of regulatory submissions to the FDA.1 High Resolution Mass Spectrometry (HRMS) is a technique often employed to confidently identify chemical components in complex mixtures but requires skilled users for operation and data interpretation. Waters recently introduced the ACQUITY RDa Detector to facilitate the deployment of accurate mass measurement technology, incorporating routine workflows and removing the need for HRMS expertise.
Here, we demonstrate the forced degradation of simvastatin under acidic, basic and oxidative conditions.2 Separation of the API (Active Pharmaceutical Ingredient) from degradants was achieved using the Waters ACQUITY UPLC I-Class PLUS.
All the major degradants were automatically identified and degradation profiles over time visualised with mass errors under 3ppm. Using full scan with both a low and high energy function, the fragmentation cone voltage was ramped, generating fragment ion information, for added structural information.
Forced degradation of pharmaceutical drug product and drug substance, as described in FDA and ICH guidelines3,4, is a vital part of pharmaceutical analytical development, requiring characterization of known and unknown impurities. While UV (Ultraviolet) detection is employed for routine monitoring of degradant products, techniques such as High Resolution Mass Spectrometry (HRMS) are required for initial compound characterisation. This technique usually requires expertise to operate the instruments and interpret the data generated.
The ACQUITY RDa Detector, with its automatic set-up and calibration, allows accurate mass measurements to be obtained by scientists with a diverse range of analytical expertise. This access to HRMS for non-expert users empowers scientists with a far greater depth of analytical information.
Additionally, the waters_connect Software platform automatically acquires, processes, and reports these results in a compliant-ready framework using dedicated end-to-end SmartMS workflows. This capability allows the scientist to use the platform easily and confidently in a regulated environment. Here, we present the forced degradation of simvastatin under separate acidic, basic and oxidative conditions, as an example of this workflow using the ACQUITY RDa Detector. We demonstrate the UNIFI Software automatically identifying and visualizing simvastatin API and the degradants generated. Using the full scan with fragmentation function, we can simultaneously acquire both high and low energy spectra, generating fragment ion information for increased confidence in compound identification.
The simvastatin standard was sourced from Sigma (Poole, Dorset). A stock solution was prepared in methanol and subjected to degradation under three separate conditions i.e. 0.5 M Hydrochloric acid, 5 mM sodium hydroxide and 10% hydrogen peroxide.
The acidic and oxidation degradation was carried out at 60 °C, and the base degradation at room temperature.
Samples were degraded over time with aliquots taken at various timepoints and analyzed to profile degradation progress.
A control solution of simvastatin in mobile phase was prepared for comparison.
|
LC system: |
ACQUITY UPLC I-Class PLUS |
|
Detection: |
TUV @238 |
|
Vials: |
TruView Max Recovery Vials, PN:186005668CV |
|
Column(s): |
ACQUITY BEH C18, 2.1 mm x 5 0mm, 1.7 µm, PN:186003554 |
|
Column temp.: |
45 °C |
|
Sample temp.: |
6 °C |
|
Injection volume: |
1 µL |
|
Flow rate: |
0.6 mL/min |
|
Mobile phase A: |
Water + 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile + 0.1% formic acid |
|
Gradient: |
25 to 90%B (8 mins) |
|
MS system: |
ACQUITY RDa Detector |
|
Ionization mode: |
ESI+ |
|
Acquisition range: |
100–2000 Da |
|
Capillary voltage: |
1.5kV (default) |
|
Cone voltage: |
20V |
|
Fragmentation cone voltage ramp: |
120–170V |
|
Scan rate: |
10Hz |
|
Desolvation gas temp: |
550 °C |
|
MS software: |
waters_connect base kit consisting of waters_connect platform, UNIFI 1.9.12 |
The ACQUITY RDa Detector set-up was fully automated, including detector set-up, auto-tune and mass calibration. Following this routine set-up, MS full scan accurate mass data was acquired at a capillary voltage of 1.5kV and a cone voltage of 20V.
Full scan mode with fragmentation function allows for cone voltage ramping to simultaneously acquire both high and low energy spectra. The automatically assigned fragmentation information provided further confidence for compound identification.
The LC/UV chromatogram was also acquired, using a Waters ACQUITY TUV @238nm, for comparison.
The simvastatin control sample was acquired with the low energy channel containing the precursor ion and the high energy channel containing the fragment ion information (Figures 1 and 2 respectively). Precursor and fragment ion assignment are automatically performed and annotated directly on the mass chromatogram.
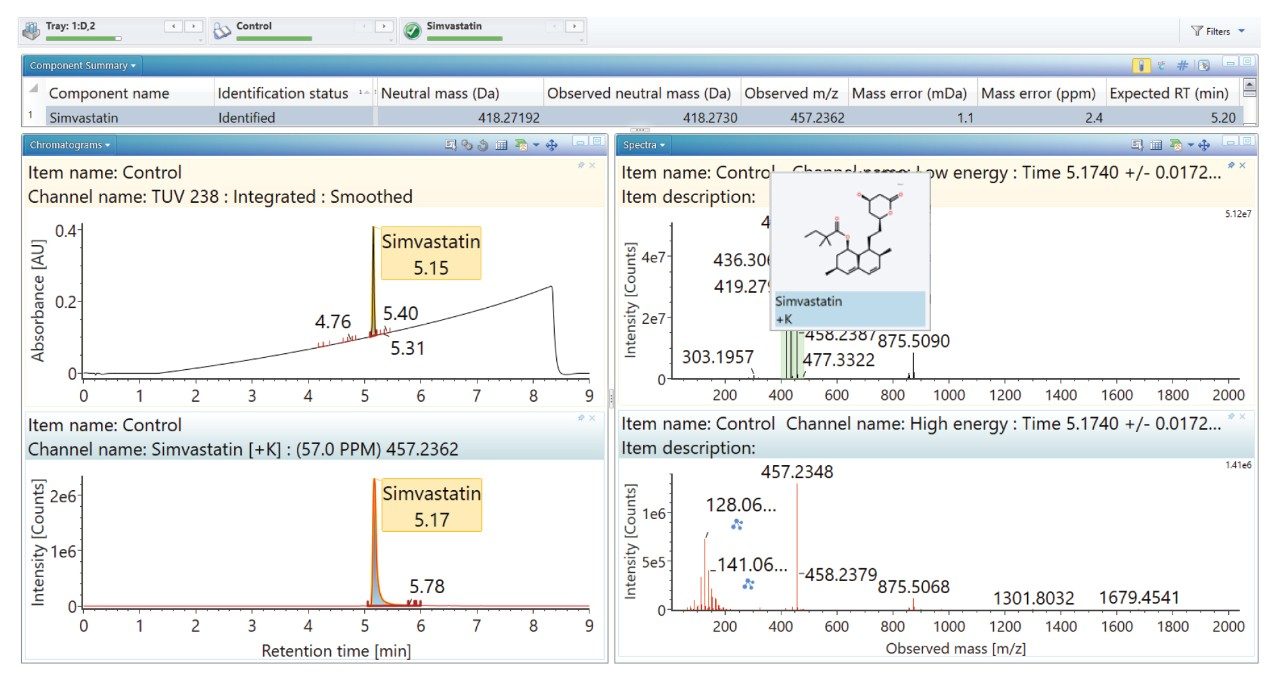
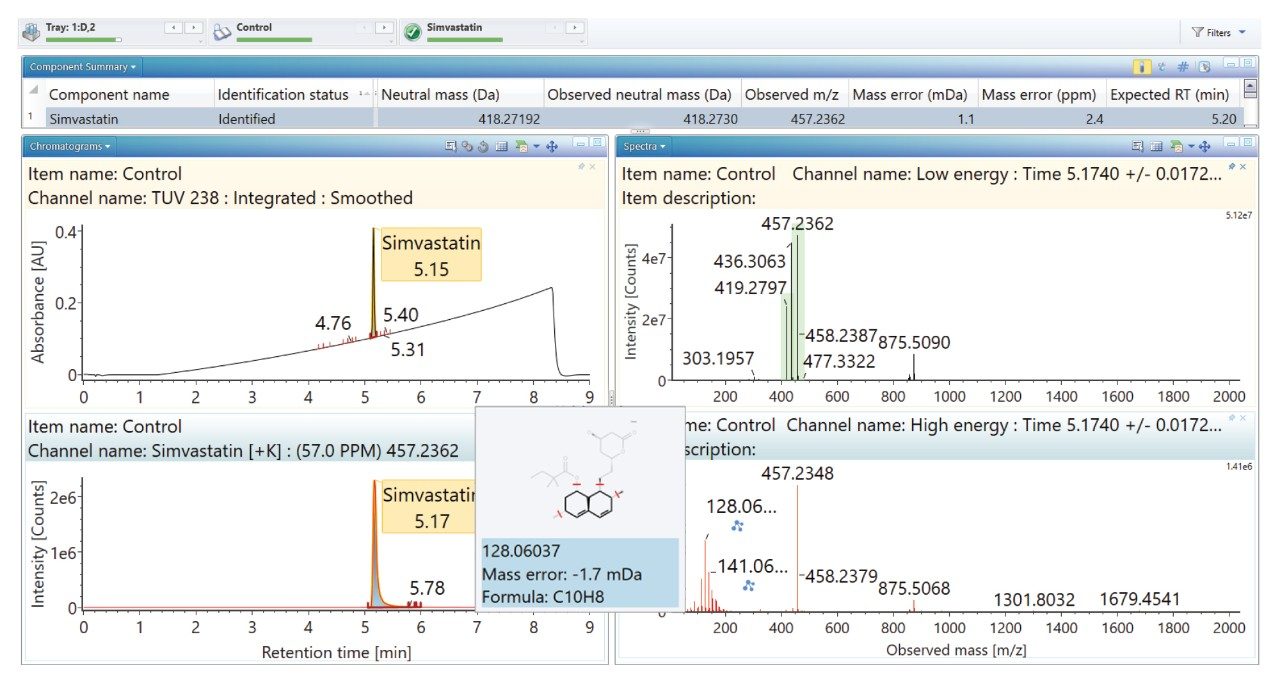
Simvastatin and its degradants readily form potassium adducts; therefore, the processing method includes [M + K]+ as well as [M + H]+ to automatically assign the detected adducts. To successfully identify degradants relating to the API, potential transformations resulting from degradation were also selected within UNIFI as part of the processing method.5,6
Selection of the ‘cleavage tool’ option enables UNIFI to automatically identify fragment ions produced via ring cleavage. This is possible due to structural knowledge of the parent molecule using chemically intelligent algorithms which perform an in silico cleavage of the molecule and identifies possible functional groups.
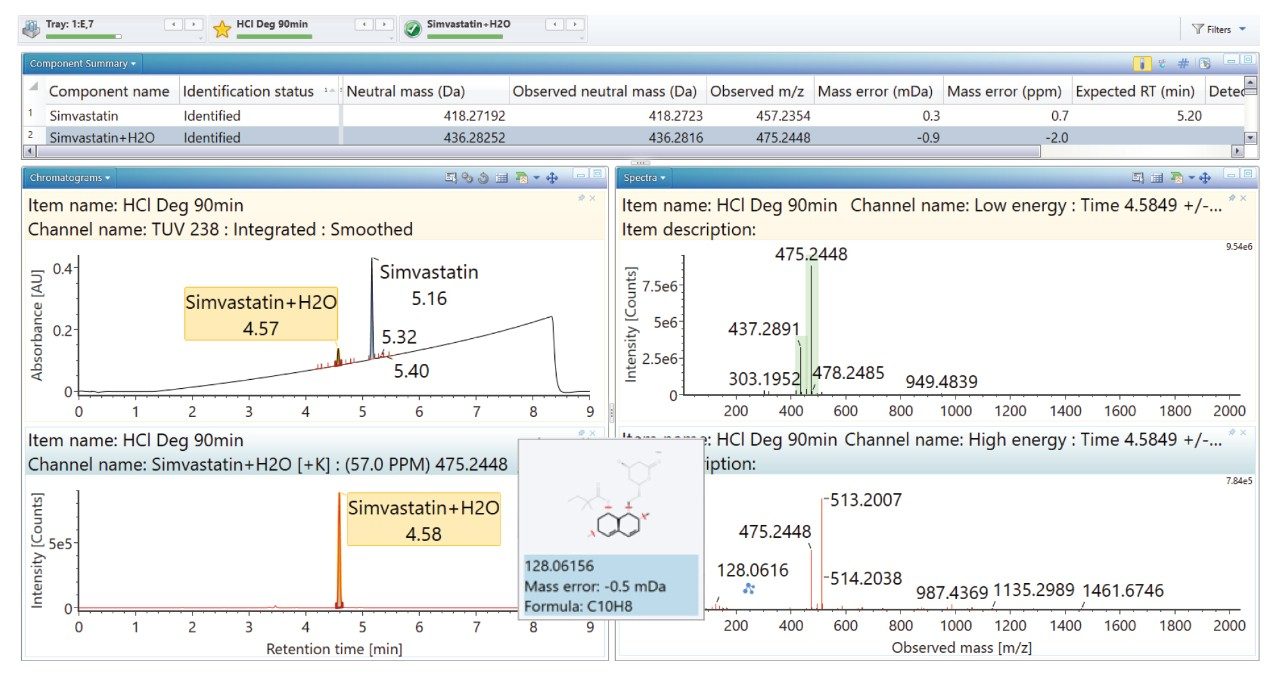
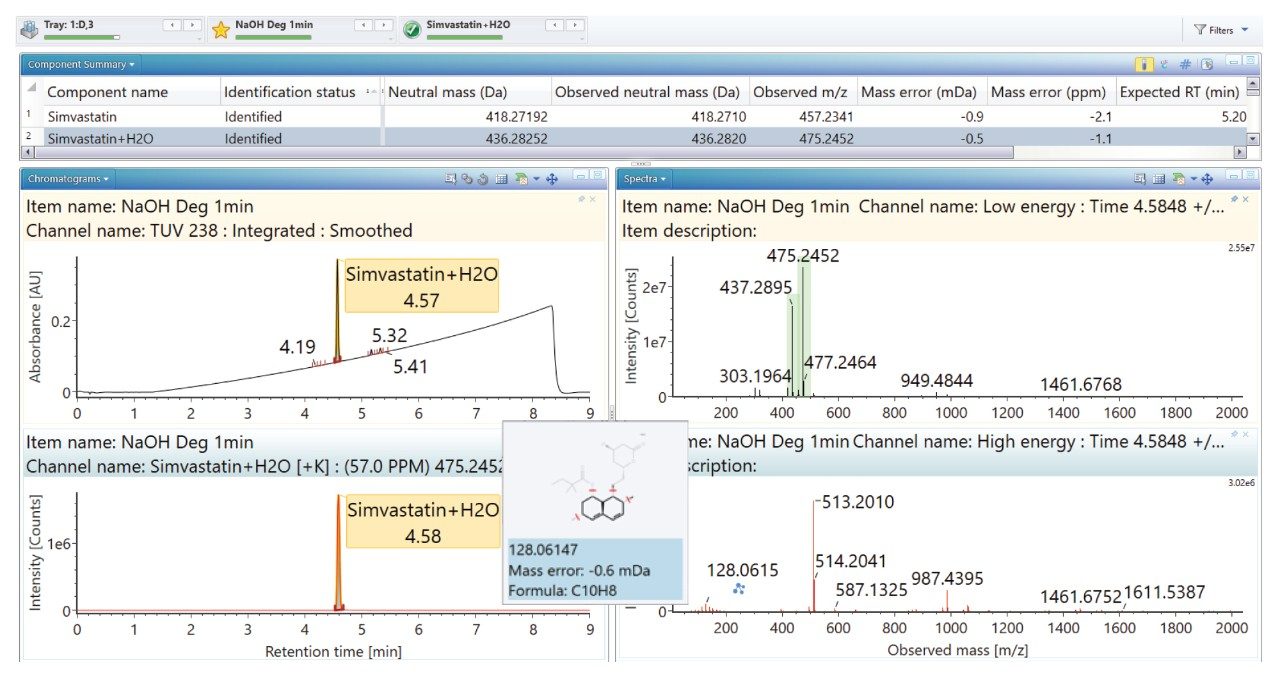
Degradation under the acidic and basic conditions described, produced a main degradant, simvastatin acid (Figure 3/4), resulting from cleavage of the tetrahydro-4-hydroxy-2H-pyran-2-one ring followed by bonding of the dihydrodiol (H2O2) group. Under oxidative conditions, the main degradant observed is the formation of the dihydrodiol species (Figure 5). All degradant structures are presented based on the API .mol File.
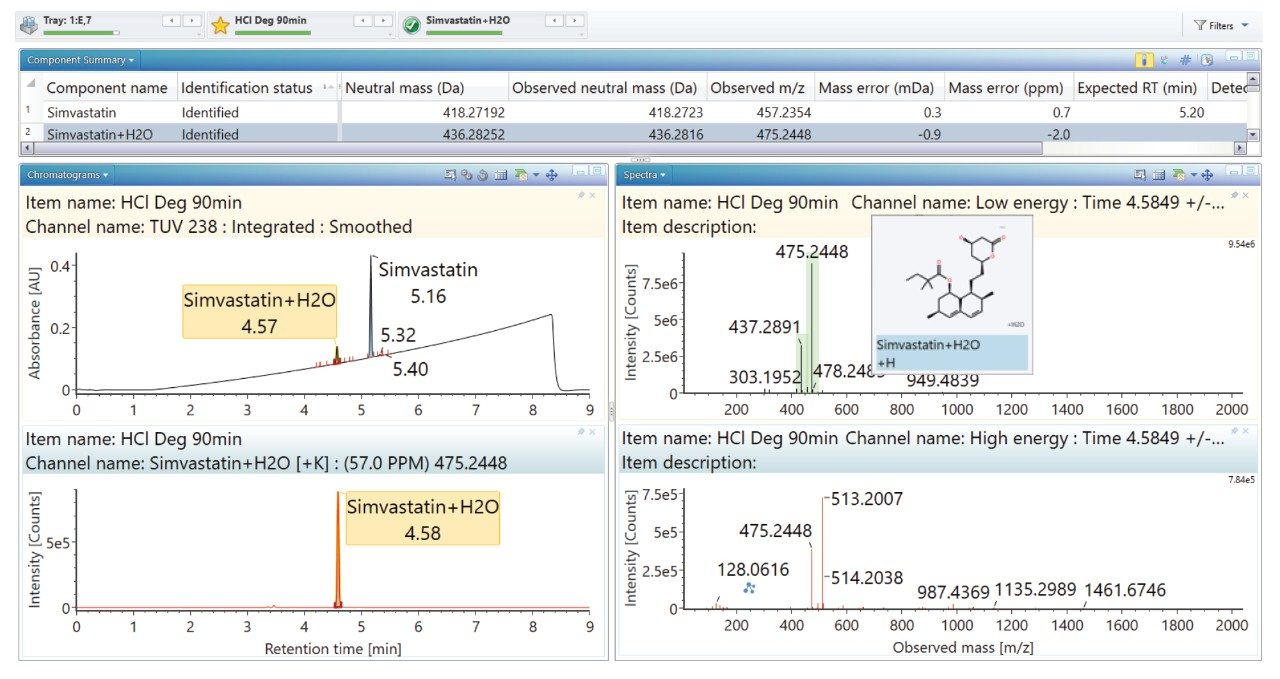
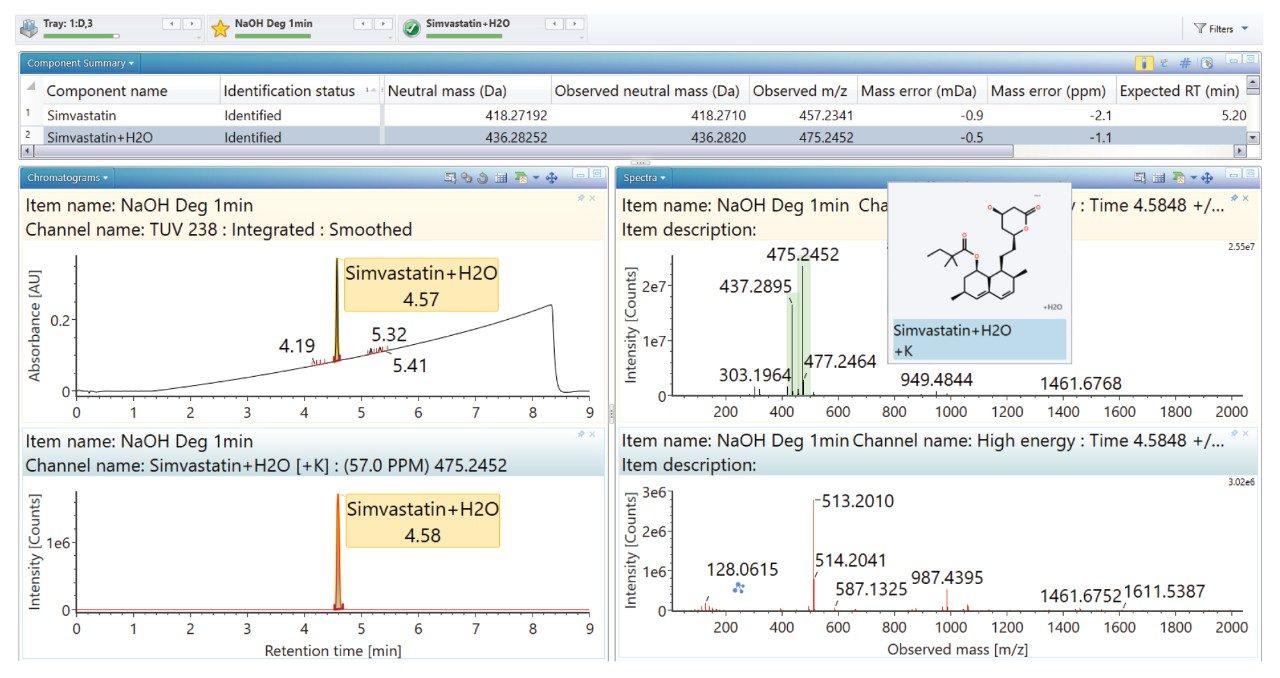
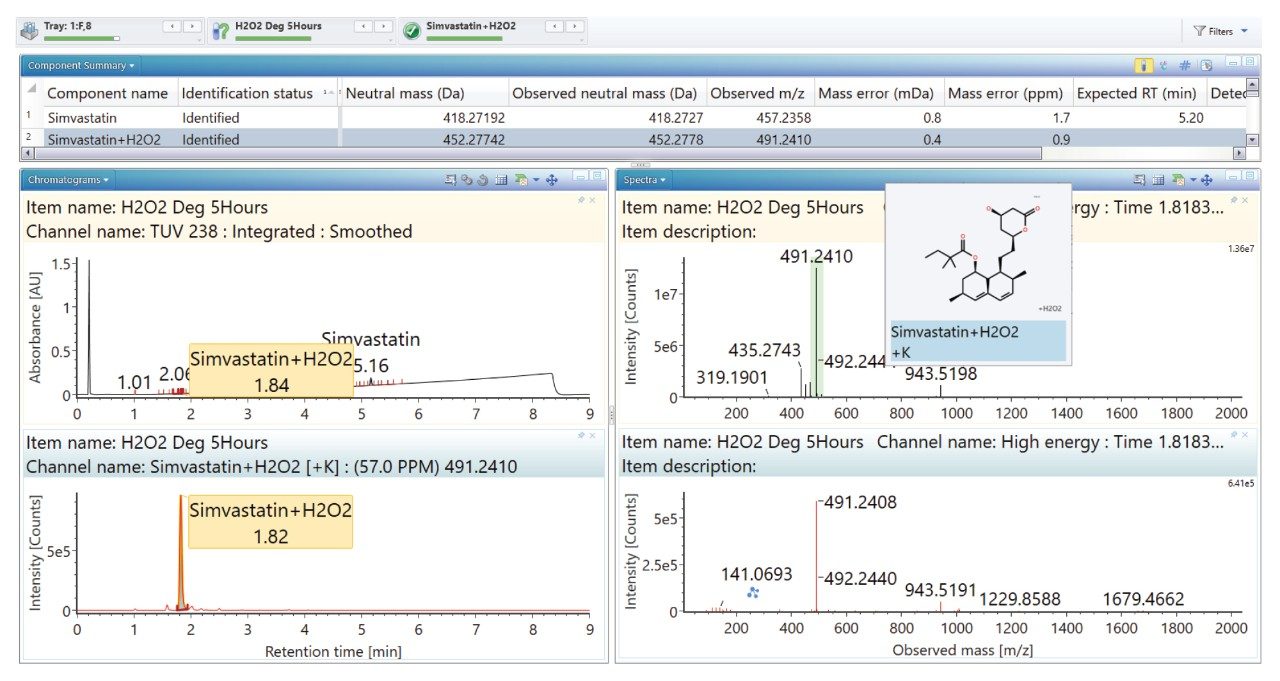
High energy spectra from cone ramping was simultaneously acquired (Figures 6, 7, and 8) with a visual representation of suggested fragmentation sites of the parent compound.
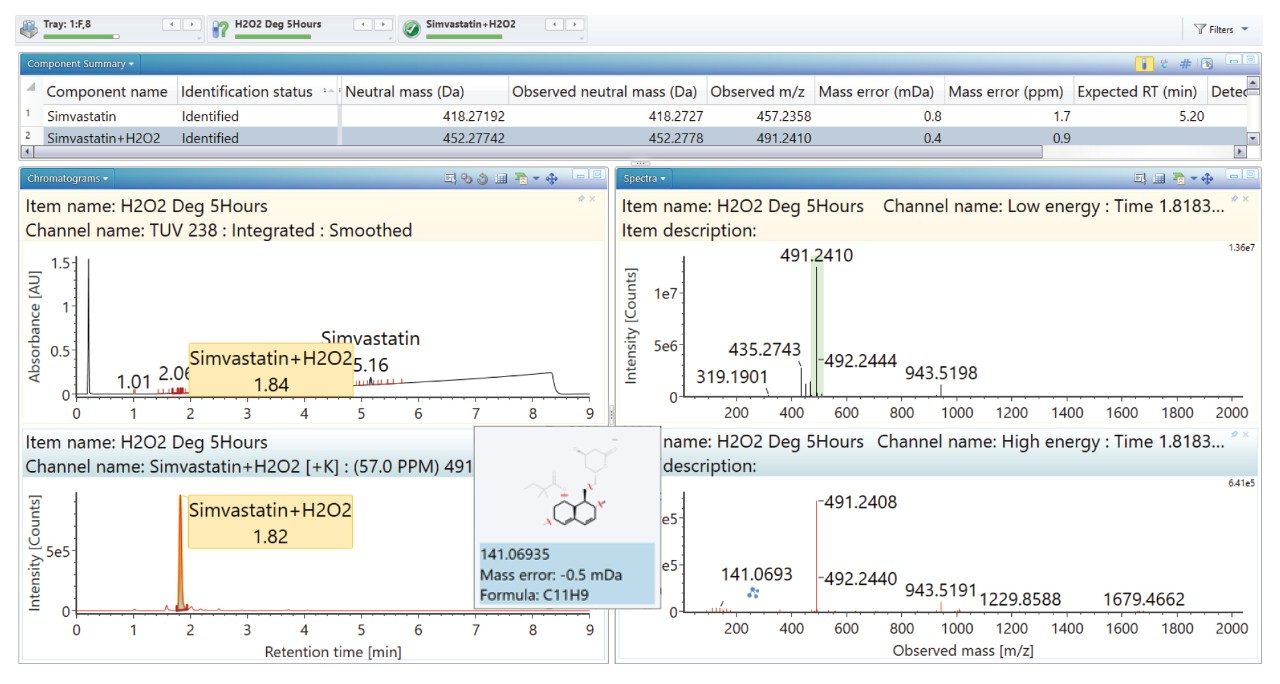
The ‘Summary Plot’ function (Figures 9, 10, and 11) provides a bar chart display showing the degradant of interest increasing over time.
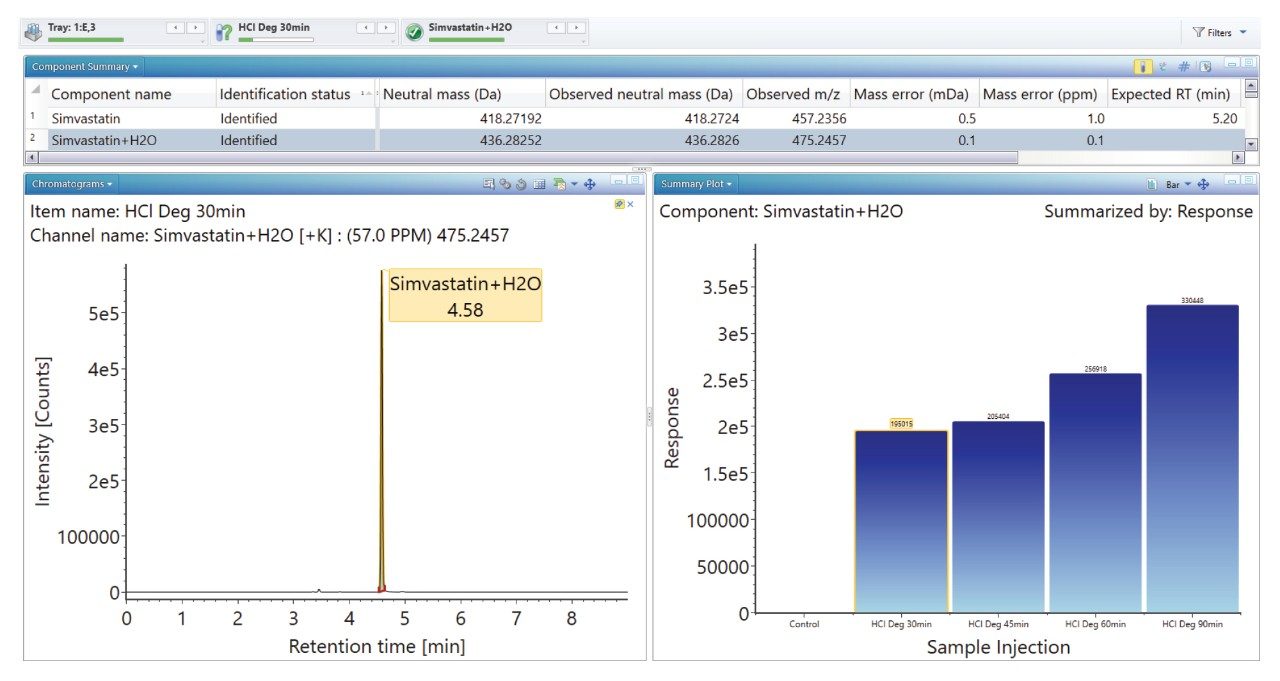
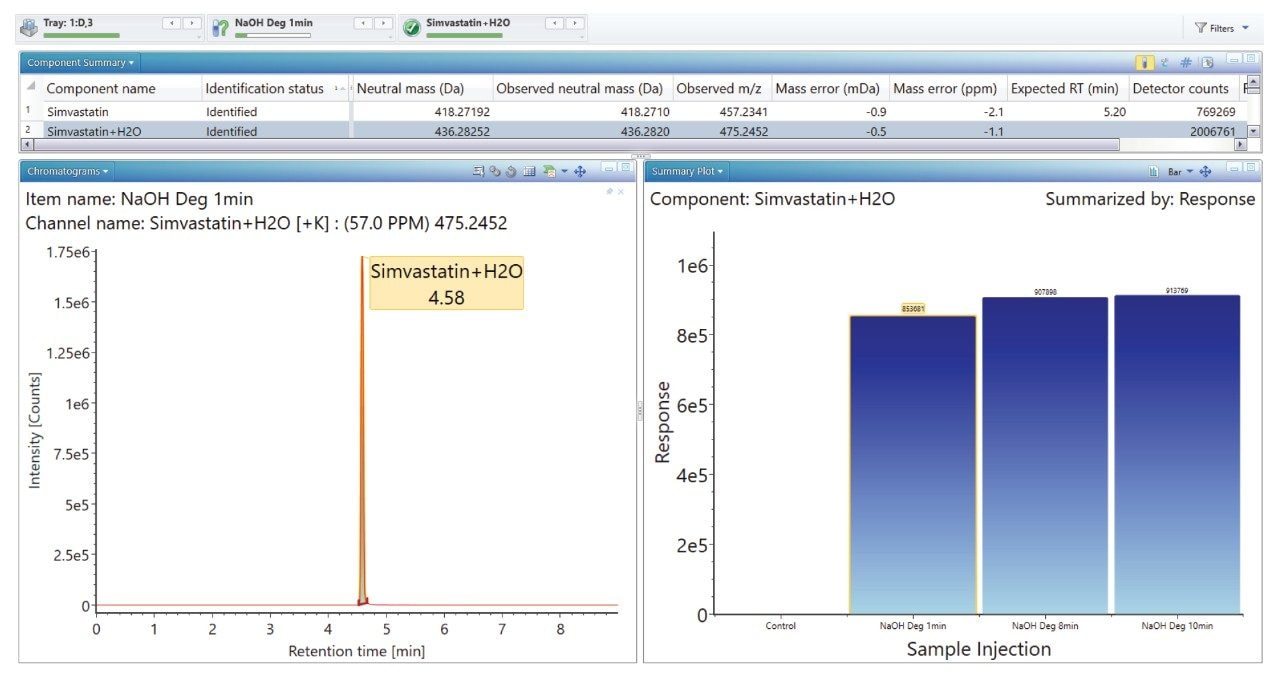
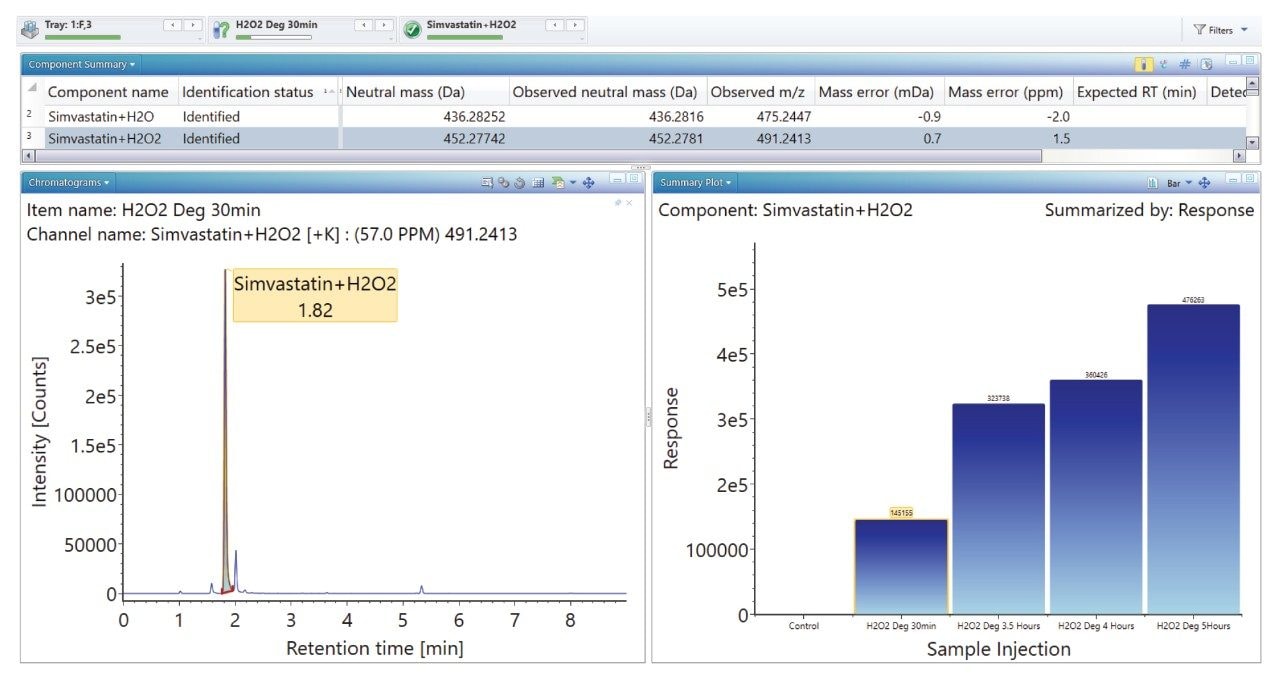
The ‘Summary Plot’ highlights that the degradation of simvastatin under basic conditions is extremely rapid with almost full conversion to simvastatin acid in under 1 minute at room temperature. Degradation of simvastatin under acidic and oxidative conditions was slower with degradation aliquots taken over 90 minutes and 5 hours respectively to achieve the desired 10-20% degradation of API.
Mass accuracy for API, degradants and fragment ions generated ranged between -1.9 and 2.4 ppm.
Forced degradation of simvastatin was successfully characterized utilizing the ACQUITY RDa Detector coupled to the ACQUITY UPLC I-Class PLUS System and TUV. The waters_connect Software platform with UNIFI application was used to automatically identify simvastatin and corresponding degradation products with a high degree of confidence. The simultaneous acquisition of high energy data, generating fragment ions, provided additional rigor to compound identification. All compounds and fragment ions presented exhibited excellent mass accuracy of less than 3ppm.
This level of compound characterization normally requires expert users in HRMS to operate the instrument and interpret the results generated. The waters_connect Software platform incorporating dedicated end-to-end workflows, combined with the ACQUITY RDa Detector’s automatic set-up, provides access to accurate mass measurement data in a routine workflow. This information rich acquisition equips the chemist with abundant information to make informed decisions on drug stability without requiring HRMS expertise.
720007124, January 2021