Streamlined LC-MS Analysis of Stress Induced Impurities of a Synthetic Peptide using the BioAccord™ System and the waters_connect™ Intact Mass™ Application
Abstract
This application note demonstrates the use of a compliance ready, automated workflow for the analysis of synthetic peptides and their impurities. The Intact Mass Application within the waters_connect informatics platform enables both detection and relative quantitation of the API (Active Product Ingredient) and its’ impurities. A noticeable benefit of the streamlined workflow is that both targeted and untargeted data processing can be achieved by adding or removing target masses within the processing method. The Intact Mass Application allows the scientist to obtain mass information, confirming the proper peptide was produced, and the relative quantitation of the detected impurities.
Benefits
- Streamlined LC-MS workflow for the mass confirmation and relative quantification of synthetic peptides and their impurities using the waters_connect™ Intact Mass Aapplication
- Capable of targeted or untargeted impurity quantification
- Built-in thresholding and pass/fail criteria ensure high data quality and the confidence to make quick assessments of the safety and efficacy of the synthetic peptide
Introduction
Peptide therapeutics are amino acid polymers containing less than one hundred amino acids.1 These are a popular class of biotherapeutics on the market, due to their relatively low toxicity, high biological selectivity, and the potential for treating a variety of diseases.2–3 Similar to other biopharmaceuticals subject to FDA approval, synthetic peptide regulatory filings require sufficient data to support its consistent manufacture, efficacy, potency, and safety. Impurities can be generated during the manufacturing process and storage.
Liquid chromatography with UV detection methods is commonly used to assess peptide impurity profiles, after those impurities are identified and risk assessed. LC-MS instrumentation and associated data processing software enables scientists to identify and quantify both the API and its impurities using increasingly simple and automated analytical workflows. This application note focuses on the capability of the recently developed waters_connect Intact Mass Application to streamline synthetic peptide analysis. The data processing workflow enables automated assignment of biomolecules based on their deconvoluted accurate mass and quantification of the components using integrated peak area from either the optical or MS data channels. This application can perform both targeted (pre-defined by mass) and untargeted analyses of analytes, enabling identity confirmation, and impurity monitoring.
To demonstrate the capabilities of the application software, we selected Exenatide as a test molecule. Exenatide is a glucagon-like-peptide-1 (GLP-1) analog, containing 39 amino acid residues. It is synthetically produced and is used in treating type II diabetes. The data shown in the application note represents a stressed Exenatide sample, a technique used in both routine and accelerated stability testing. The data were collected using the BioAccord™ LC-MS system, also operated by waters_connect operating system. This allows the full workflow automation from data acquisition to reporting of results to happen within a single compliant-ready and network-scalable informatics platform. These results highlight the capability and functionality of waters_connect and the Intact Mass Application that can be used for synthetic peptide analysis across the full product life cycle of a synthetic peptide.
Experimental
Sample Description
Stress sample preparation: Exenatide peptide was purchased from the USP (The United States Pharmacopeial Convention, Rockville, MD). The sample was dissolved in 3.0 mL of milli-Q water to make a 0.9 mg/mL solution. The concentration was adjusted to 0.5 mg/mL with pH 7.5 100 mM Tris buffer to obtain six aliquots of the diluted sample. Three aliquoted samples were covered to prevent exposure to light and incubated at 37 °C for two to four days. The second set of aliquots were incubated at room temperature, shielded from ambient light, for the same amount of time. At the end of the incubation period both samples were diluted in 0.1% formic acid to a final concentration of 0.1 mg/mL for LC-MS analysis. The on-column injection volume was 1 µL.
Exenatide Sequence Information
HGEGTFTSDL SKQMEEEAVR LFIEWLKNGG PSSGAPPPS-NH2
Monoisotopic mass: 4184.02731 Da
Average mass: 4186.57188 Da
LC-MS system suitability analysis: MassPREP™ Peptide mixture (p/n: 186002337) was dissolved in 100.0 µL of 0.1% formic acid. Injection volume was 1 µL. The peptides selected for monitoring and the neutral mass information are listed below.
|
Peptide name: |
Bradykinin, Angiotensin II, angiotensin I, Renin substrate, Enolase T35 |
|
Molecular weight: |
1059.5613,1045.5345,1295.6775,1757.9253,1871.9604 Da |
|
Relative quantification: |
Peak area/total response/exclude raw |
|
LC minimum peak area: |
5% |
|
Pass/warning/fail thresholding for impurities: |
5%/2%/1% |
|
Iintensity threshold (minimum): |
10% (of the largest peak) |
Method Conditions
|
LC Conditions |
|
|
LC system: |
ACQUITY™ Premier System |
|
Detection: |
TUV (214 nm), MS |
|
Vials: |
QuanRecovery with MaxPeak™ HPS (p/n: 186009186) |
|
Column(s): |
ACQUITY Premier Peptide CSH C18 Column 1.7 µm, 130 Å, 2.1 x 100 mm (p/n: 186009488) |
|
Column temp.: |
60 °C |
|
Sample temp.: |
6 °C |
|
Injection volume: |
5 µL |
|
Flow rate: |
0.25 mL/min |
|
Mobile phase A: |
0.1% formic acid in H2O |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
|
Gradient: |
10% B for 2 min, 95% B for 13 min, 85% B for 2 min, 90% B for 6 min |
Gradient Table

MS Conditions
|
MS system: |
BioAccord System with ACQUITY Premier |
|
Ionization mode: |
ESI+ (MS with fragmentation) |
|
Acquisition range: |
m/z 50–2000 |
|
Capillary voltage: |
1.2 kV |
|
Collision energy: |
60–120 V |
|
Cone voltage: |
20 V |
Informatics Tools
|
Data processing application: |
Intact Mass App v1.2 |
|
Informatics platform: |
waters_connect v2.0 |
Selected Intact Mass App Processing Parameters
|
Deconvolution method: |
Auto |
|
Maximum number of peaks to deconvolute: |
10 |
|
LC minimum peak area: |
1 |
|
1–15 KDa algorithm: |
BayesSpray |
|
Output mass: |
Monoisotopic |
|
Type of biomolecule: |
Protein |
|
Relative quantitation: |
Area, largest response, exclude raw |
|
Specify intensity threshold (minimum): |
0.1% most intense peak |
Results and Discussion
LC-MS data was acquired with reversed-phase separation prior to MS acquisition using the BioAccord system (ACQUITY UPLC-ToF MS). Both the UPLC and the columns employ MaxPeak High Performance Surfaces (HPS) technology to minimize non-specific interactions between peptides and the surface of the fluidic path. ACQUITY Premier Reverse Phase Columns have shown to improve acidic peptide recovery resulting in the reduced need for column pre-conditioning and low variability between replicate measurements.4,5 The samples were separated using a 15-minute gradient on an ACQUITY Premier Peptide CSH C18 Column.
It was reported that Exenatide is susceptible to chemical degradation by pH variance, heat treatment, and light exposure.6 To generate a sample containing known impurities associated with thermal degradation process, half the aliquots of the Exenatide sample were incubated at elevated temperature for two to four days at pH 7.5. At the end of incubation period the samples were subjected to LC-MS analysis. The total ion chromatogram (TIC) of the heat stressed sample is shown as Figure 1A. The extracted region of the chromatogram, on the top left, shows the main peak of Exenatide and several potential degradation products (1–5).
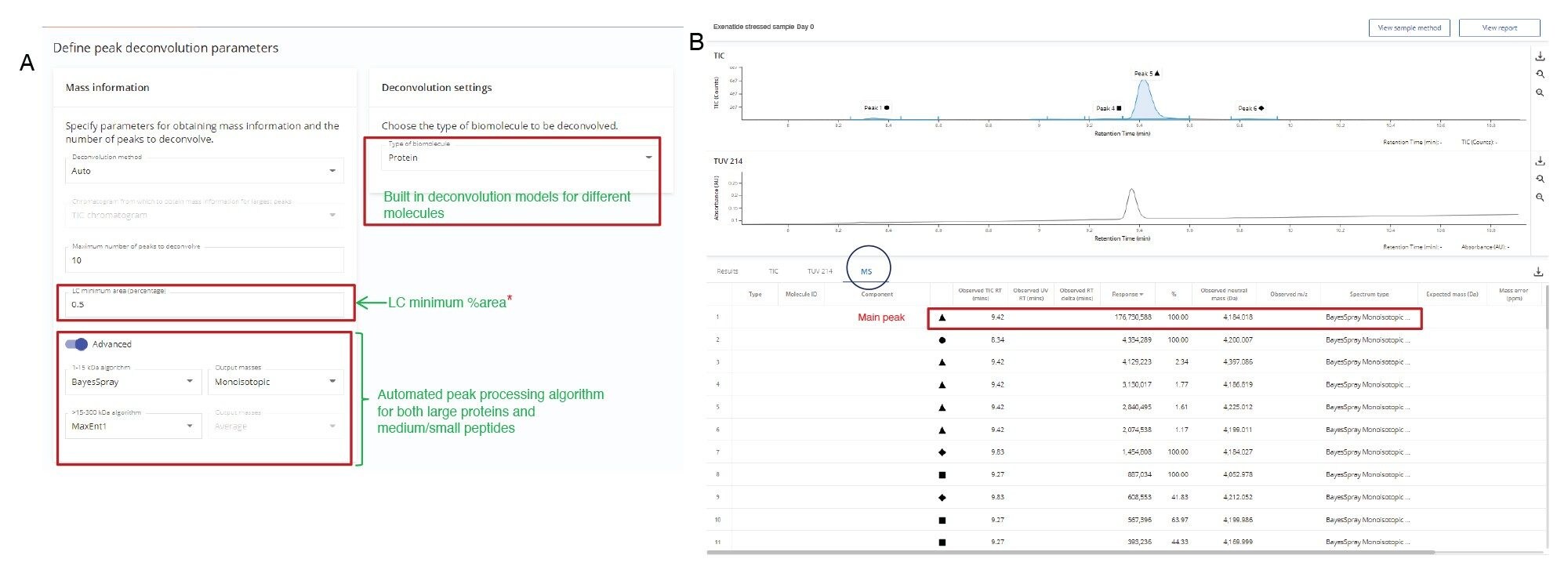
An untargeted analysis (with no searched components based on mass) was performed on the initial Exenatide LC-MS data to assign the peaks for the API and its impurities, as shown in Figure 1B. In the analysis method, the expected masses were assigned to each injection to establish individual target masses for subsequent analyses. Each summed mass spectra for peaks present above 0.5% peak area in the TIC (calculated relative to the total peak area of all the peaks) was charge deconvoluted to obtain neutral mass information. The Intact Mass Application can automatically adjust charge deconvolute settings based on the m/z profiles of analytes (e.g., isotopically resolved, and unresolved regions of spectra). The charge deconvolution settings used for Exenatide are listed under “biomolecule” in the “Define Deconvolution Parameters” section of the method (Figure 2A) using a refined isotopic model for “Protein” molecules. The BayesSpray algorithm was selected to provide monoisotopic deconvolution results for molecules with mass between 1–15,000 Da, since the synthetic Exenatide has a MW at 4184.0273 Da.
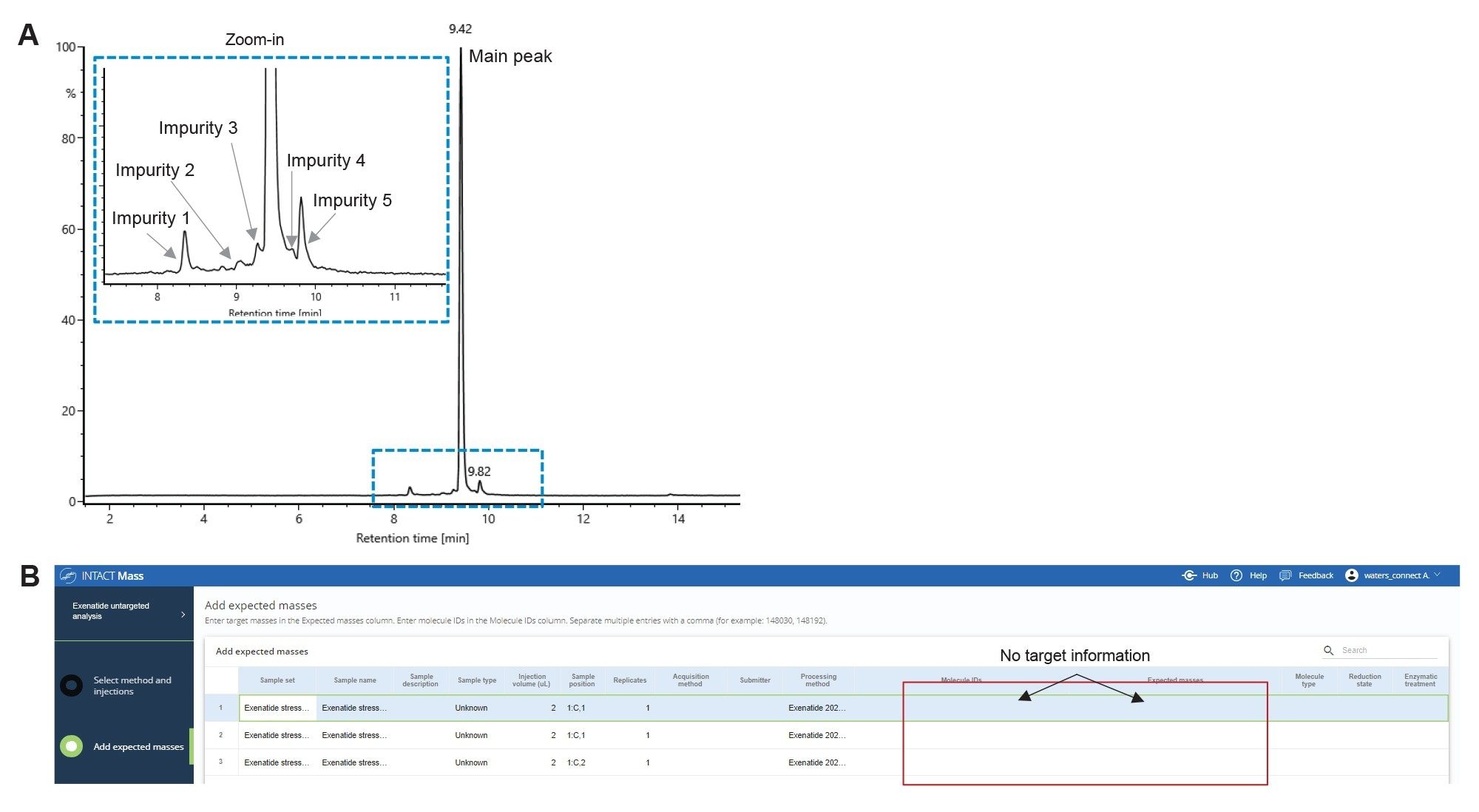
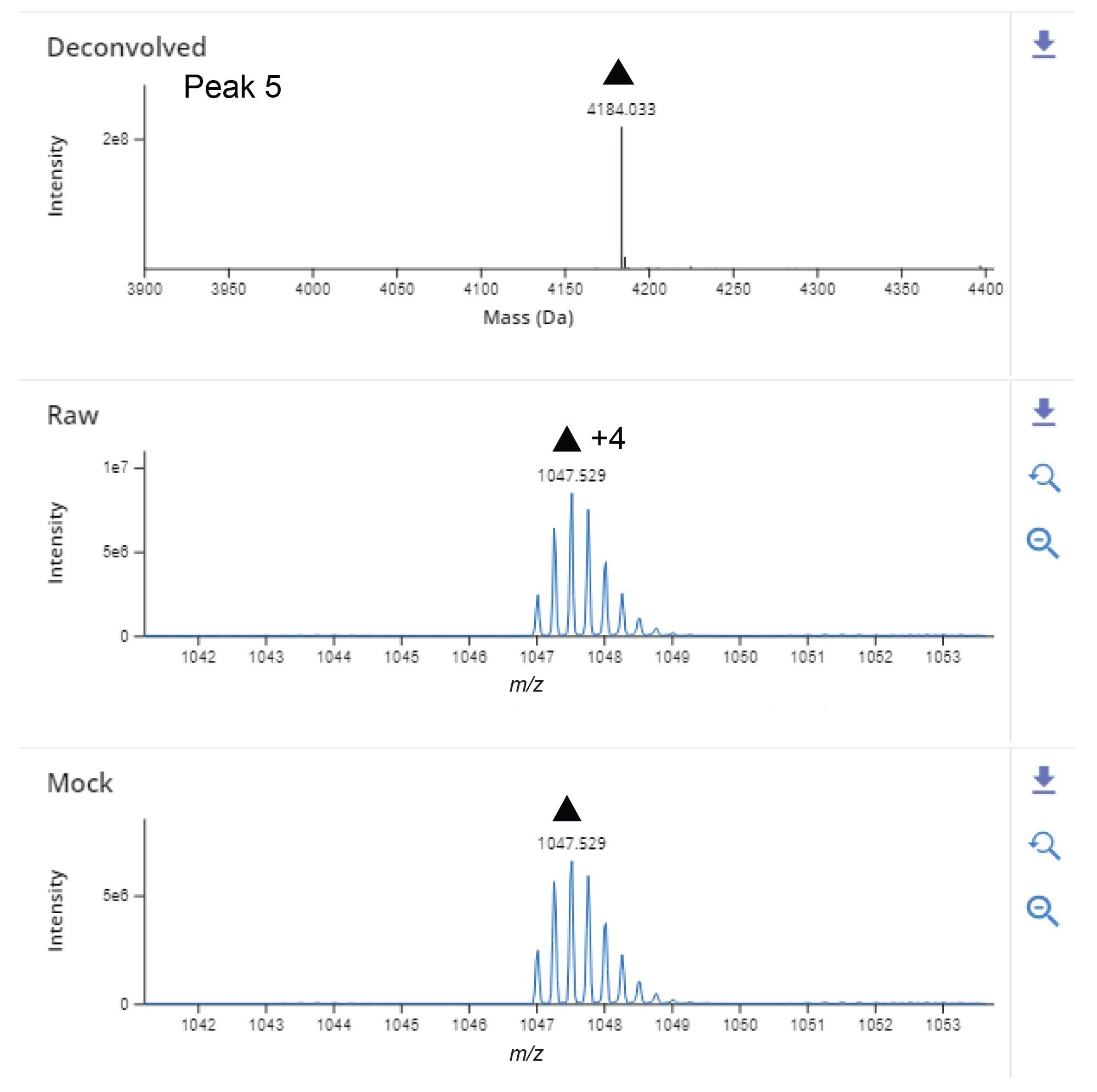
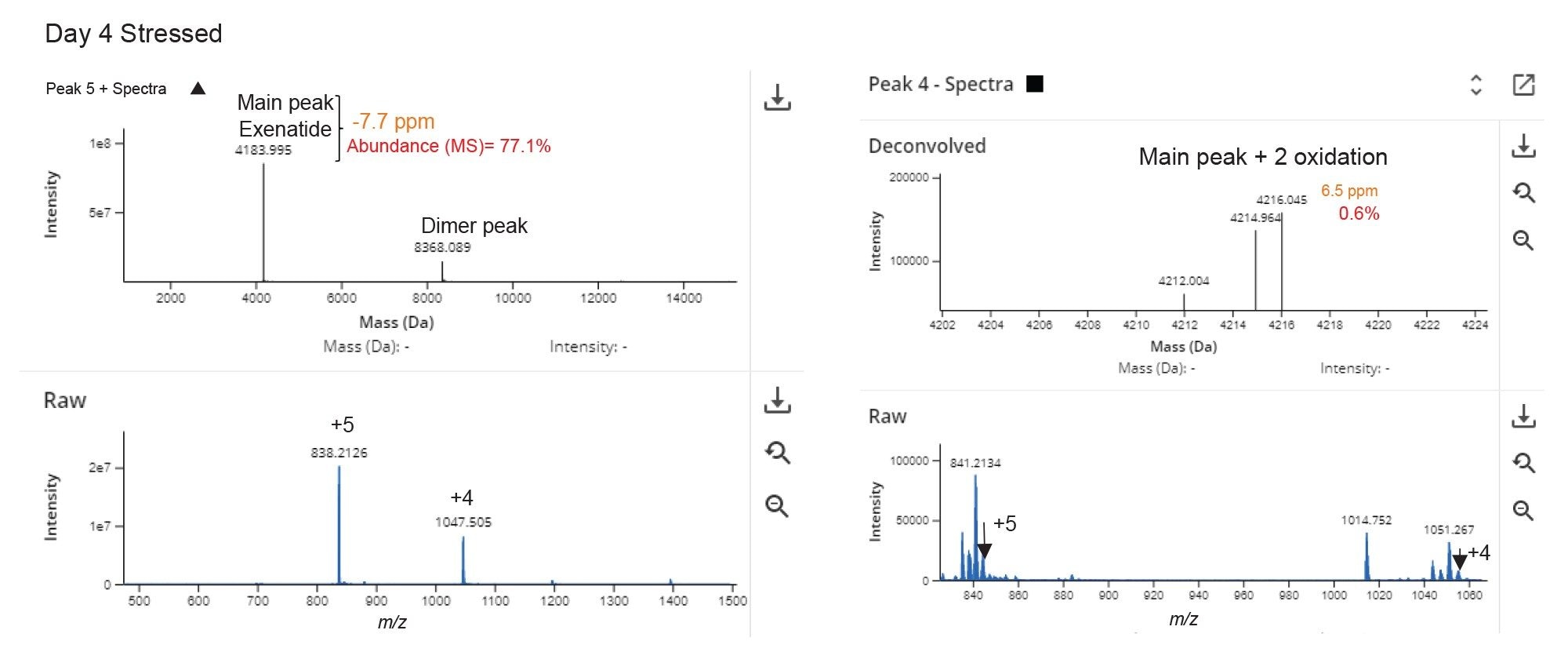
Figure 2B shows the total ion chromatogram (TIC) for the Exenatide peptide with Peak 5 as the most intense chromatographic peak eluting at 9.42 minutes. The deconvoluted data identified the neutral mass of the base peak as 4184.018 Da with 1.7E8 MS response. The main charge states observed for this peak were +4 and +5 (Figure 3). Figure 3 shows the data for the largest peak from the untargeted analysis matched the theoretical monoisotopic mass of Exenatide within 5 ppm mass accuracy (mass error=-2.2 ppm).
In addition to mass confirmation of the main API peak, the untargeted analysis offers a straightforward way to assign the masses to unknown impurity peaks in the chromatogram. These impurities can then be specified in a targeted analysis (Figure 4A), that matches the mass of the unknown peak to an Exenatide mass containing a single or multiple modifiers. The modifiers used in the case study were determined based on the mass shift (delta mass from the API) calculated from untargeted analysis data. The list of modifications included potential impurities related to synthesis process and peptide degradation due to stress conditions (data from the day 4 stressed sample). These modifications are oxidation, deamidation, deletion of methionine (desMet), succinimide modification of asparagine (Asn), and aggregate/dimer formation (Figure 4B). The neutral mass of exenatide was added in as an additional modifier to target the impurity peak due to aggregation multimeric peaks of exenatide, a common impurity produced due to heat stress.6 As shown in Figure 4B, each modification was clearly labeled to confirm the impurity status. An embedded scientific library in the Intact Mass Aapplication is typically used to generate the modifier list. The impurities identified in the stress sample (day 2 heat stress) are noted in Figure 5. Based on the results, the most abundant impurities were oxidation, deamidation succinimide modification, and loss of methionine. Each modification, independent of relative abundance, was identified with ≤10 ppm mass error. The data reported here are based on TIC, however, both TIC and optical (UV) chromatograms can be used in peak identification and quantification.
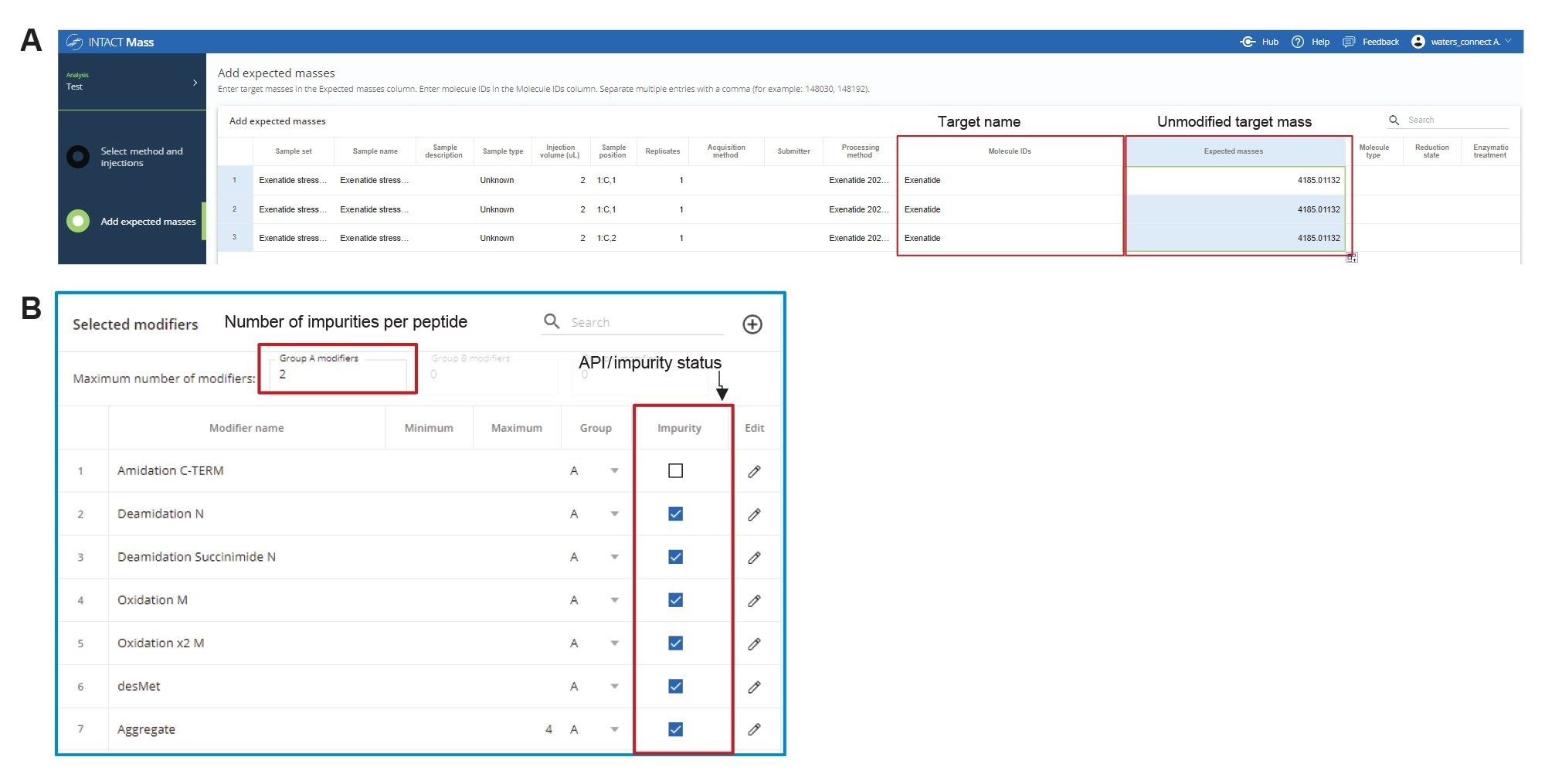
B) Each injection is set up as untargeted analysis.
API Purity and Impurity Quantifications
API Impurity assignment is often followed by purity assessment of the Drug Product (DP). The peptide purity level was determined by the following equation: percent peptide purity = [(Area UV 220nm DP)/ (Area UV 220nm all peaks)] *100; where: Area UV 220 UV nm DP is the peak area for the desired peptide in the LC chromatogram monitored at 220 nm, and Area UV 220 nm all peaks is the sum of the areas for all peaks. Following the same formula for optical detection, the example data was used for purity calculation using MS response (TIC) as well. Figure 5 shows the percent purity of the stressed sample (four days at elevated temperature). In the purity calculation, the C-terminal amidation was grouped together with native Exenatide MS response to obtain the total percent purity of the drug product. The C-terminal amidation is a common modification observed in synthetic peptides that improves the stability of the peptide.
Like API purity, the Intact Mass Application calculates relative quantification for impurities using the peak area from TIC or UV trace. Further, the percent %abundance of an impurity can be calculated based on peak area or peak height relative to the total response of all the peaks or the most abundant peak. The Day 4 stressed impurities were calculated using the peak area of impurity relative to the peak area of the most abundant peak. A pass/fail criterion was implemented to demonstrate a built-in quality control feature. We reported the quantitation value using TIC due to low UV signal under the formic acid mobile phase conditions. Using lower levels of TFA as the ion pairing reagent would be beneficial for obtaining sensitive UV signals, should the quantitation using UV trace is preferred, but would require higher sample loadings to overcome TFA induced electrospray signal suppression.
The most recent guidelines published by FDA for synthetic peptide drugs require the relative quantification of impurities that are more than 0.5% abundance of the drug substance/API.7 Therefore, in this case study, 0.5% was selected as the minimum quantification threshold. In the example data, only peaks at or more than 0.5% abundance were reported. The most abundant impurity was the aggregate (dimer) of Exenatide which was measured at 16.8% abundance (day 2 stressed sample), and completely absent in the reference sample (day 0) confirming the susceptibility of Exenatide to elevated pH and heat effects. Other major impurities were oxidation at 4.7%, loss of methionine (desMet) at 1.4% and asparagine succinimide at 5.9% relative to the main peak.
The result table in Figure 5, displays a variety of other measurements: such as product purity, modifier assigned, peak retention time, MW, mass error, quantitation, and pass/fail status. The purity of the Exenatide stressed sample was flagged due to the API purity which was below the expected value set at 95%. The lowest reported impurity (0.8%) identified in the Day 4 stressed sample had a double oxidation peak of Exenatide with 6.5 ppm mass accuracy (Figure 6). The abundance of this peak relative to the total MS response of all the identified peaks using deconvoluted MS spectra was ~0.6%, showing high quality results for both major and minor detected impurities.

Conclusion
- An automated LC-MS workflow for synthetic peptide mass confirmation and impurity profiling was accomplished using the BioAccord LC-MS system within the waters_connect informatics platform
- Stressed Exenatide peptide was used to show the process of API identity confirmation, purity assessment, and impurity definition and quantitation. Quality features such as System Suitability Test, selectable thresholding, and pass/ fail criteria were illustrated
- The utility to perform an initial untargeted impurity characterization for initial peak assignments and library generation, followed by subsequent targeted monitoring was shown to offer the flexibility and usability for analysts to evaluate the product quality attributes, and rapidly evolve methods with new product knowledge
References
- FDA; 2016b Guidance for Industry: Biosimilars: Questions and Answers Regarding Implementation of the Biologics Price Competition and Innovation Act of 2009; 2016.
- Mason, J. M., Design and development of peptides and peptide mimetics as antagonists for therapeutic intervention. Future Medicinal Chemistry 2010, 2 (12), 1813–1822.
- Pernot, M.; Vanderesse, R.; Frochot, C.; Guillemin, F.; Barberi-Heyob, M., Stability of peptides and therapeutic success in cancer. Expert Opinion on Drug Metabolism & Toxicology 2011, 7 (7), 793–802.
- Robert E. Birdsall, Jacob Kellett, Samantha Ippoliti, Nilini Ranbaduge, Matthew A. Lauber, Ying Qing Yu, Weibin Chen, Reducing Metal-ion Mediated Adsorption of Acidic Peptides in RPLC-Based Assays Using Hybrid Silica Chromatographic Surfaces, Journal of Chromatography B, 2021, 122700, ISSN 1570–0232
- Ranbaduge, N., Birdsall, R., Yu, Y.Q, Chen, W., The BioAccord system with ACQUITY Premier for improved peptide monitoring, Waters literature, 720007351, August 2021.
- Benet A, Halseth T, Kang J, Kim A, Ackermann R, Srinivasan S, Schwendeman S, Schwendeman A. The Effects of pH and Excipients on Exenatide Stability in Solution. Pharmaceutics. 2021 Aug 16;13(8):1263. doi: 10.3390/pharmaceutics13081263. PMID: 34452224; PMCID: PMC8398870.
- FDA ANDAs for Certain Highly Purified Synthetic Peptide Drug Products That Refer to Listed Drugs of rDNA Origin; Food and Drug Administration: https:\\www.regulations.gov, 2017.
720007752, October 2022