Targeted and Non-Targeted Identification and Characterization of the Forced Degradation Products of Glipizide Using the ACQUITY RDa Detector and UNIFI Software Workflows
Abstract
In this application note we demonstrate the use of the ACQUITY RDa Detector to provide routine access to high resolution mass spectrometry (HRMS) data for the characterization of products generated from the forced degradation of glipizide.
A sample of glipizide was subjected to chemical stress under acidic, basic, and oxidative conditions. The separation of the drug and its degradation products was carried out on an ACQUITY UPLC I-Class PLUS Binary System. The degraded samples were analyzed using the ACQUITY RDa Detector, a benchtop TOF mass analyzer, coupled to an ACQUITY PDA (Photodiode Array) Detector.
The workflow is separated into two phases, which represent targeted and non-targeted approaches (Figure 1). In the first phase we demonstrate the screening of known degradants taken from the literature1 (labelled Impurities I-V). In the second phase, plausible degradations are generated from the structure of the Active Pharmaceutical Ingredient (API).
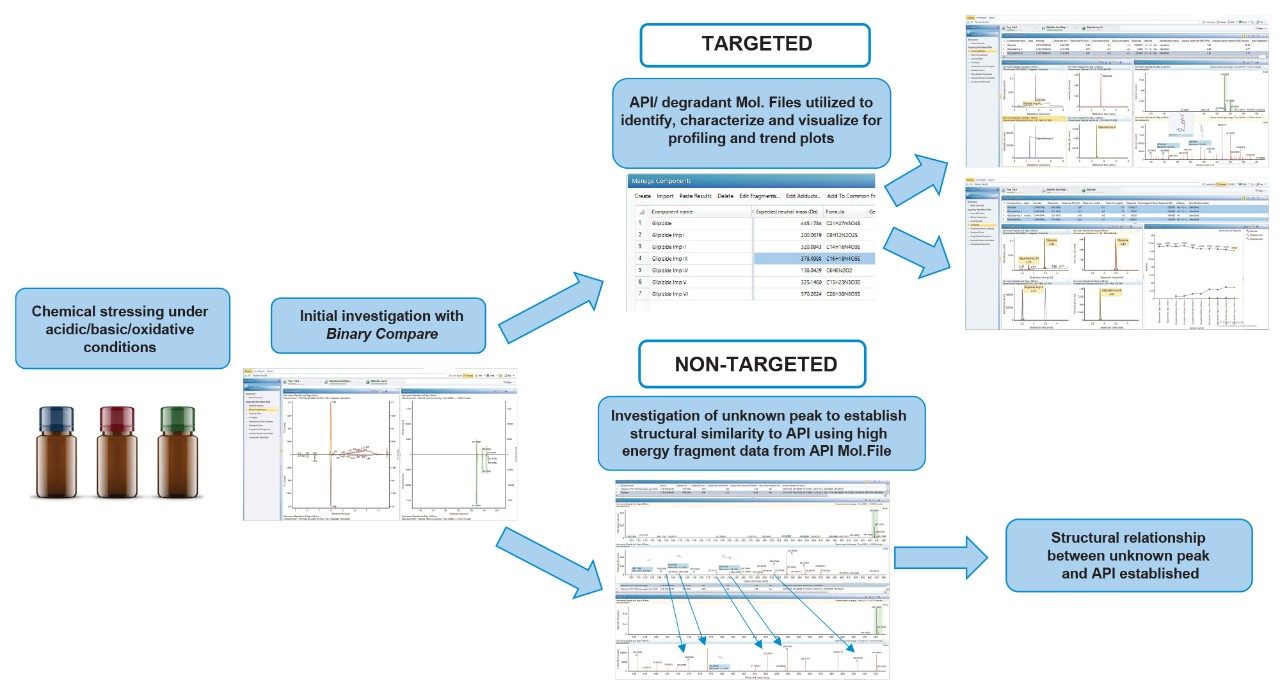
In the targeted workflow phase, glipizide was found to produce impurities II and III under acidic and oxidative conditions with the latter also generating the oxidation product detected at m/z 478.1746 tentatively assigned by UNIFI as [M + O2]+. Impurity V was generated exclusively under basic conditions. Identification and characterization of the main compound, the known impurities and the associated fragments generated, was carried out automatically using libraries within the processing method.
For the non-targeted workflow, confirmation of the identity of the unknown oxidation product was sought through fragmentation. Structural information of the suspected degradant was gained by comparing high energy fragments of glipizide, based on the structural file (.mol) and the high energy fragment ions generated by the identified impurity in the oxidatively stressed sample. Fragment libraries were created using the high energy data of the API and imported into the processing method. The acquired data were screened against a method containing the parent compound and potential transformations, thereby identifying, and visualizing any related structures. Using this approach the software was able to confidently assign the unknown impurity at m/z 478.1746 as [M+ O2]+ .
Here we demonstrate a routine, streamlined workflow for both the targeted and non-targeted characterizing of forced degradation studies. This routine access to accurate mass data, enables compound characterization without the need for HRMS expertise, and unknown peaks can be identified without requiring sample submission samples to a dedicated HRMS lab.
Benefits
- Routine access to accurate mass measurements for targeted and non-targeted forced degradation studies for a range of MS expertise
- Automatic identification and visualization of degradant and fragment ions without the need for manual interpretation
- Visualization of potential structural fragment ion detection to aid in pathway profiling studies
- Streamlined workflows within UNIFI allowed for a stepwise investigation of the data allowing quick appraisal of degradants produced to detailed profiling of degradants over time
- waters_connect platform allows for a fully 21 CFR Part 11 compliant end-to-end workflow creation for routine analysis of forced degradation
Introduction
The forced degradation study is considered a vital analytical aspect of the drug development program for pharmaceuticals.
Knowledge of the degradation pathways of a drug and gaining an insight into any degradation products through characterization using for example HRMS is vital in understanding the intrinsic stability of a molecule.
As per International Conference on Harmonization (ICH) guidelines (Q1A), stability studies need to be performed to propose the shelf life of new drug substances and/or drug products. Shelf-life studies are part of various regulatory submissions to the U.S. Food and Drug Administration (FDA).2,3
Knowledge of the stability of molecule helps in selecting proper formulation and package as well as providing proper storage conditions and shelf life, which is essential for regulatory documentation. Forced degradation is a process that involves degradation of drug products and drug substances at conditions more severe than accelerated conditions and thus generates degradation products that can be studied to determine the stability of the molecule.4
The capabilities of the analytical procedures used to assess the effects of stress testing are fundamental to their ability to provide assurance that the pharmaceutical product meets applicable standards of identity, strength, quality, and purity during its shelf life. Within this application note we detail the forced degradation of glipizide, a second-generation anti-diabetic sulfonylurea drug under acidic, basic, and oxidative conditions and present the findings from both a targeted and non-targeted workflow approach.
Experimental
Sample Description
A standard of glipizide (Sigma, Poole, Dorset) was prepared at a concentration of 1 mg/mL in 80:20 methanol:water. Next, 900 µL aliquots of this solution were added to amber vials and chemically stressed with 100 µL of 99% formic acid, 50% NaOH, and 30% hydrogen peroxide (six preparations under each stress condition). Each vial was incubated at 80 °C over timepoints t= 0, 30, 60, 120, 180, and 240 minutes. After these timepoints have elapsed, a sample under condition was removed from the heat and cooled. A 100 µL aliquot of each stressed sample was taken and diluted 1:10 with 80:20 methanol:water before being submitted for analysis.
LC Conditions
|
LC system: |
ACQUITY UPLC I-Class PLUS |
|
Detection: |
ACQUITY PDA (Photodiode Array) at 254 nm |
|
Vials: |
TruView Max Recovery Vials, (p/n: 186005668CV) |
|
Column(s): |
ACQUITY UPLC BEH C18 100 x 2.1mm, 1.7µm (p/n: 186002352) |
|
Column temp.: |
45 °C |
|
Sample temp.: |
8 °C |
|
Injection volume: |
1 µL |
|
Flow rate: |
0.4 mL/min |
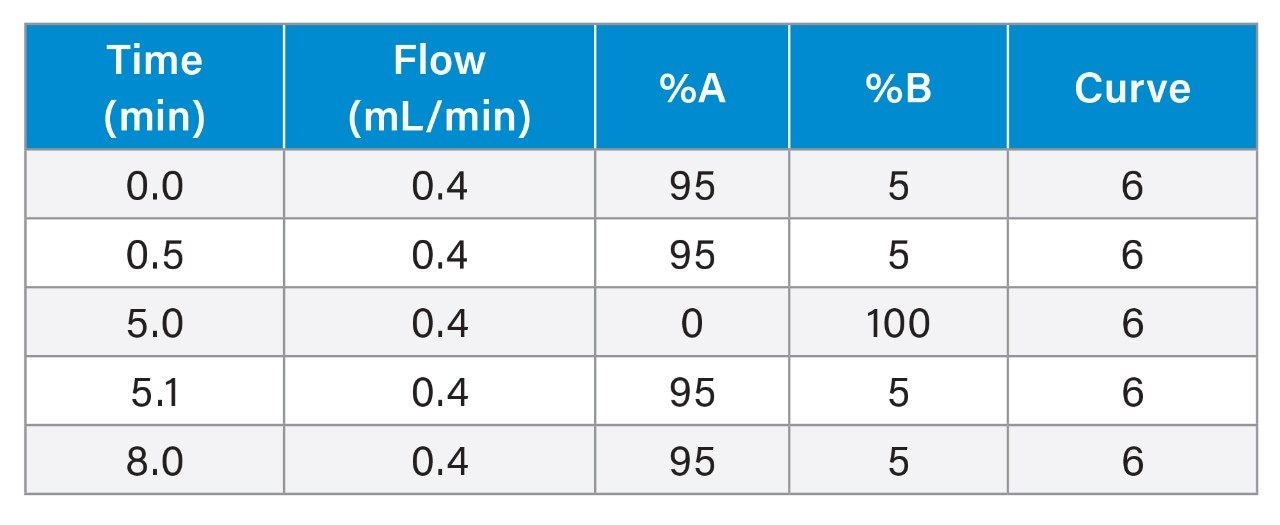
Gradient Table
MS Conditions
|
MS system: |
ACQUITY RDa Detector |
|
|
Ionization mode: |
Positive |
|
|
Acquisition range: |
50–2000 m/z |
|
|
Capillary voltage: |
1.5 kV (default) |
|
|
Fragmentation cone voltage: |
60 V–150 V |
|
|
Cone voltage: |
30 V |
|
Data Management
|
MS software: |
UNIFI 1.9.13.9 |
|
|
Informatics: |
waters_connect |
|
Results and Discussion
The ACQUITY RDa Detector was set up automatically, including detector, auto-tune, and mass calibration. Following this routine set up, MS full scan accurate mass data were acquired at a capillary voltage of 1.5 kV and a cone voltage of 30 V. Using the Full Scan with Fragmentation function allows cone voltage ramping to simultaneously acquire high and low energy spectra. The high energy data function, containing fragment ion information, was assigned automatically providing further confidence for compound identification and degradation pathway profiling of the parent drug.
In the targeted phase, glipizide was found to produce impurities II and III under acidic and oxidative conditions, with the latter also generating a suspected oxidation product at m/z 478.1746 not cited previously in literature for glipizide decomposition studies. Impurity V was generated exclusively under basic conditions.
Structures for the non-targeted analysis were generated based on the structure of the parent drug with predicted/potential transformations included in the processing method. Selection of the cleavage tool option enables UNIFI to automatically identify fragment ions produced via bond cleavages. This is possible due to structural knowledge of the parent molecule using chemically intelligent algorithms which perform an in silico cleavages. To further assist in confirming a structural relationship between the parent drug and degradants produced, fragments generated from the API were added to the processing method and screened for as ‘expected fragments’.
Targeted Analysis
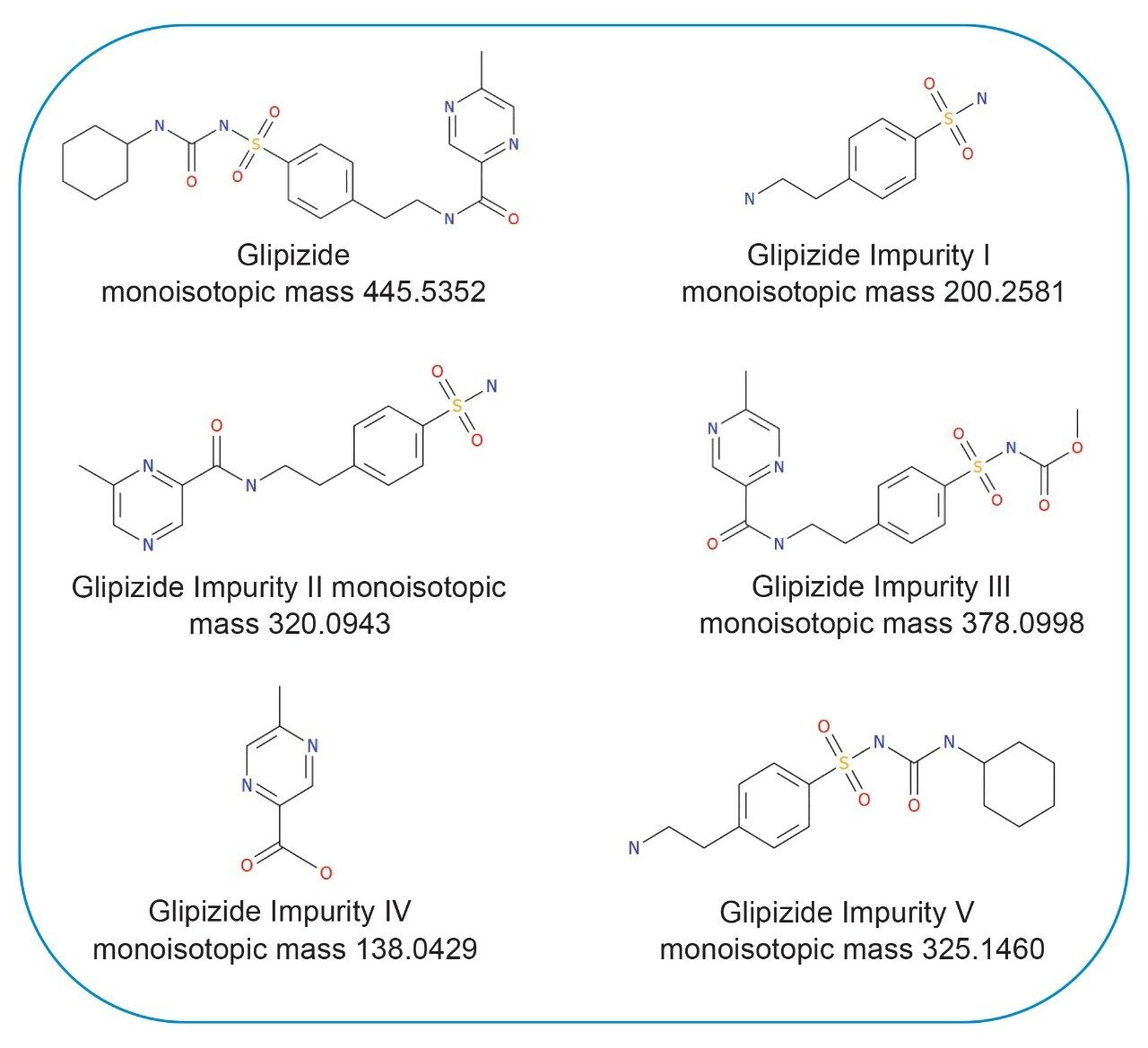
Within the UNIFI application a targeted workflow was created using a library based on the structures of known degradants and use that structural information to screen the data collected for any degradants that have been formed during the incubations. Here, the screening library consisted of five known impurities (Figure 2). UNIFI uses this library directly on the detected component table to search for known degradants.
Binary Compare
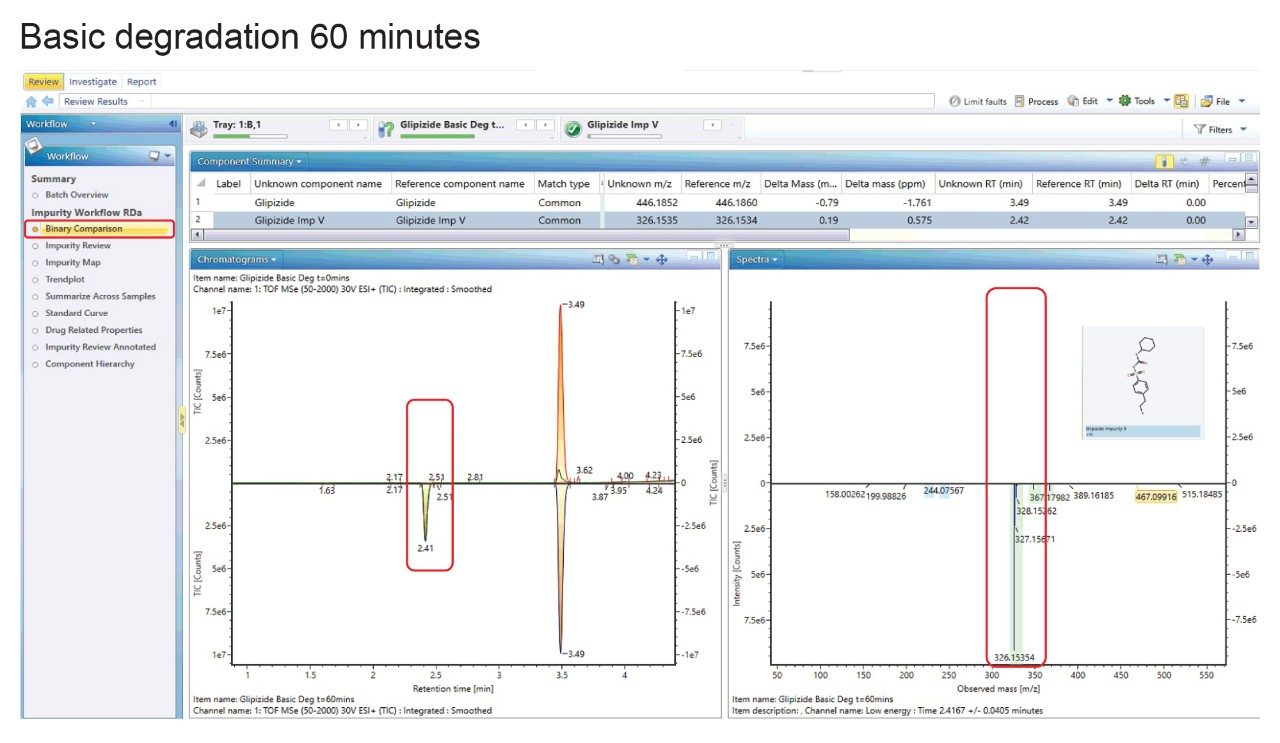
The Binary Compare function was employed for an initial appraisal of the incubated samples. This takes a reference sample, in this case we have the basic degradation t=0 minutes, and an unknown, with the basic degradation t=60 minutes selected (Figure 3). A peak at 2.41 minutes is visible in the degraded sample only, and selection of that peak gives a spectral peak of m/z 326.1534 which UNIFI identifies as glipizide impurity V. The structure is also visualized from the information contained in the structural file (.mol).
Impurity profile
Further interrogation of the data under all conditions was carried out by comparing the high and low energy channels generated using the full scan with fragmentation function.
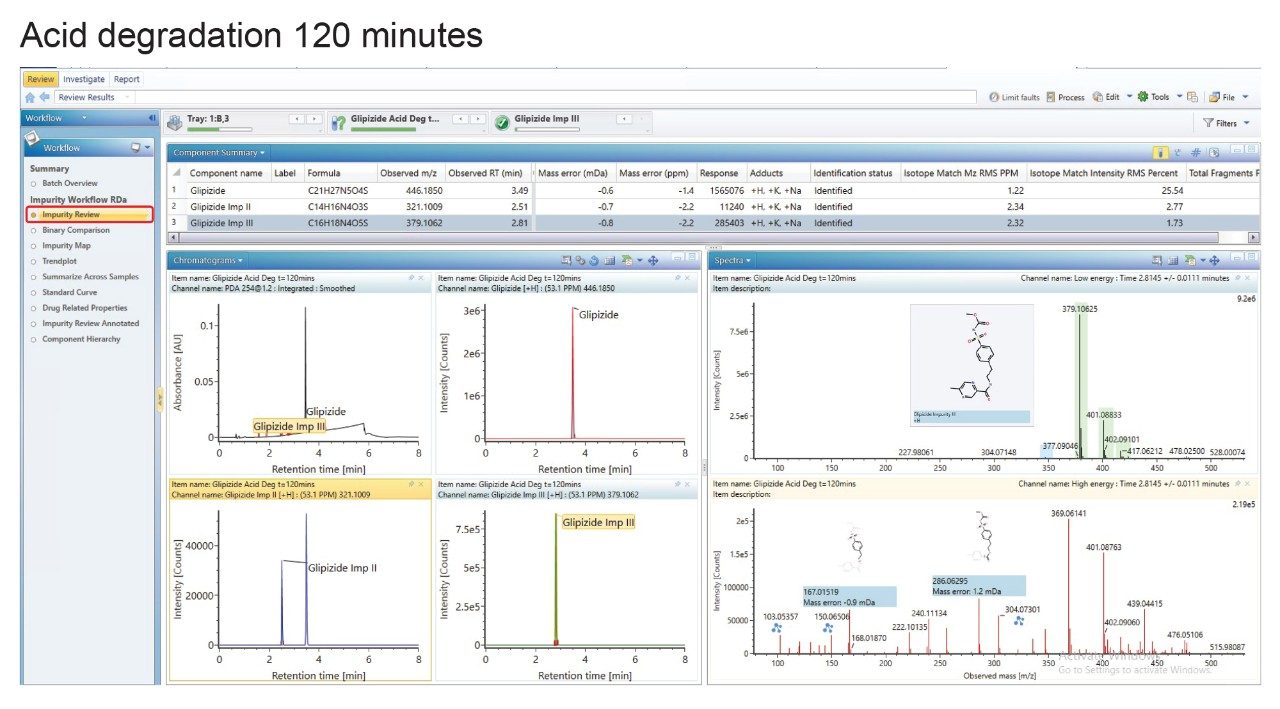
As an example, the acid degradation sample at t=120 minutes, which has resulted in the generation of impurities II and III - both cited previously in literature – identified automatically by the software based on the structural file (.mol) information.
As shown in Figure 4, the low energy channel, the selected product ion (impurity III) has been assigned and visualized automatically, with the high energy channel showing the fragmentation profile of the impurity. Multiple fragment ions have been detected with the of fragment ions at m/z 167.0151 and m/z 286.0629 visualized, and the charge-retaining portion highlighted.
Trend plot
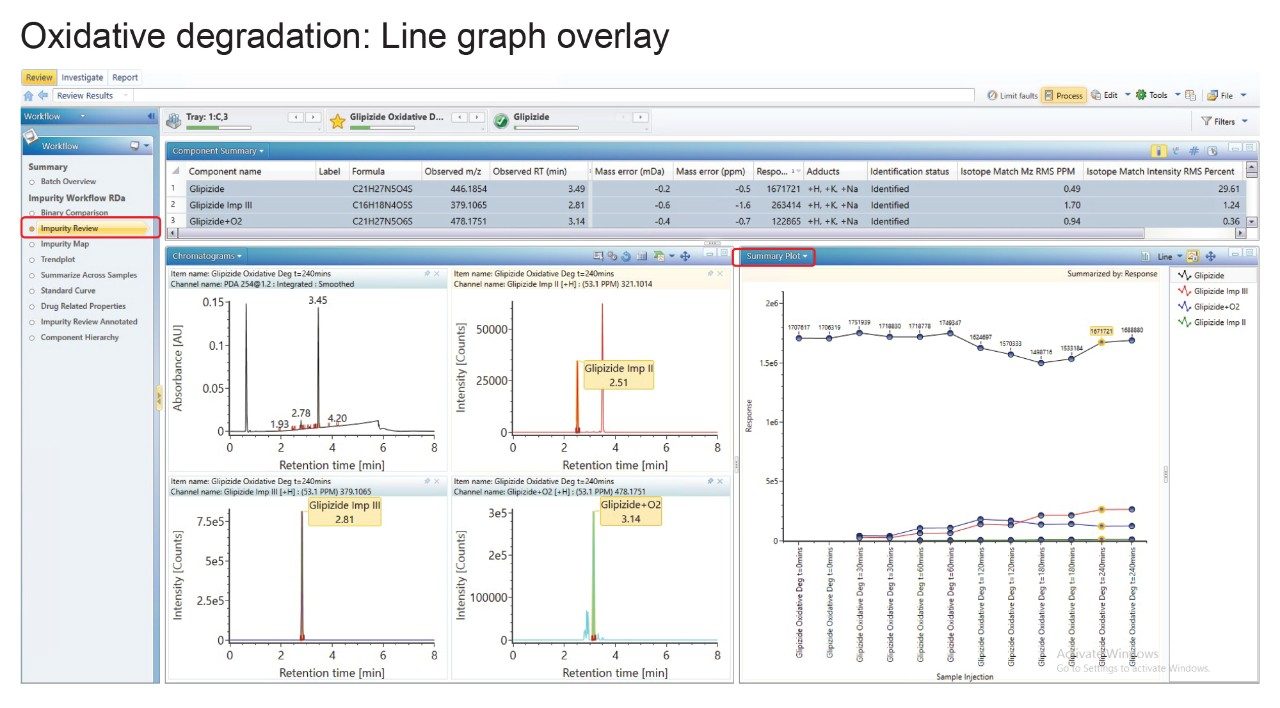
Using the oxidation incubation as an example (Figure 5), the line graph demonstrates the decrease in glipizide response and concomitant increase in impurities II and III as well as the oxidation product at m/z 478.1751. The slight increase in response of the glipizide at 240 minutes scan be attributed to evaporation due to high percentage of organic solvent at elevated temperatures.
Trend plot across all stress conditions
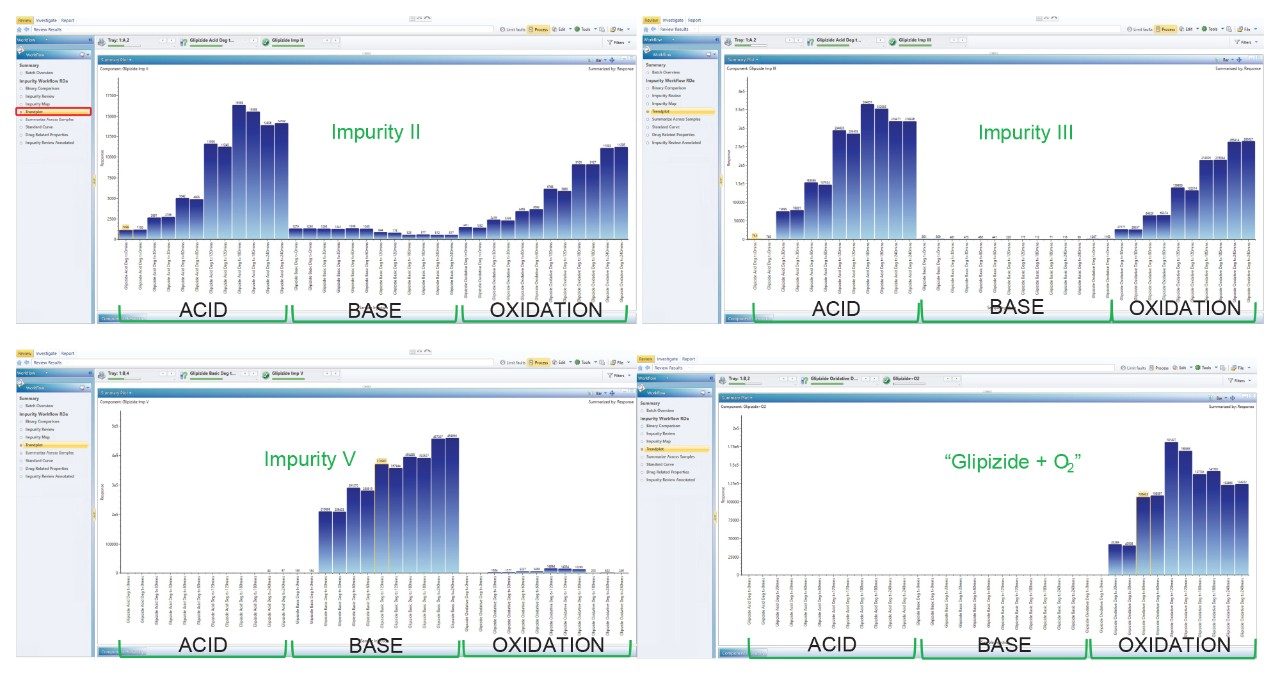
The Trend plot workflow step was modified to show the spread of impurity generation over all the timepoints tested, giving an indication of glipizide relative stability under all the stress conditions used (Figure 6). As can clearly be seen from these graphs, impurity II and III are generated under predominantly acidic and oxidative conditions, impurity V exclusively under basic conditions and the oxidation product at m/z 478.1746 described in the non-targeted analysis section, generated under oxidative conditions only.
Non-Targeted Analysis
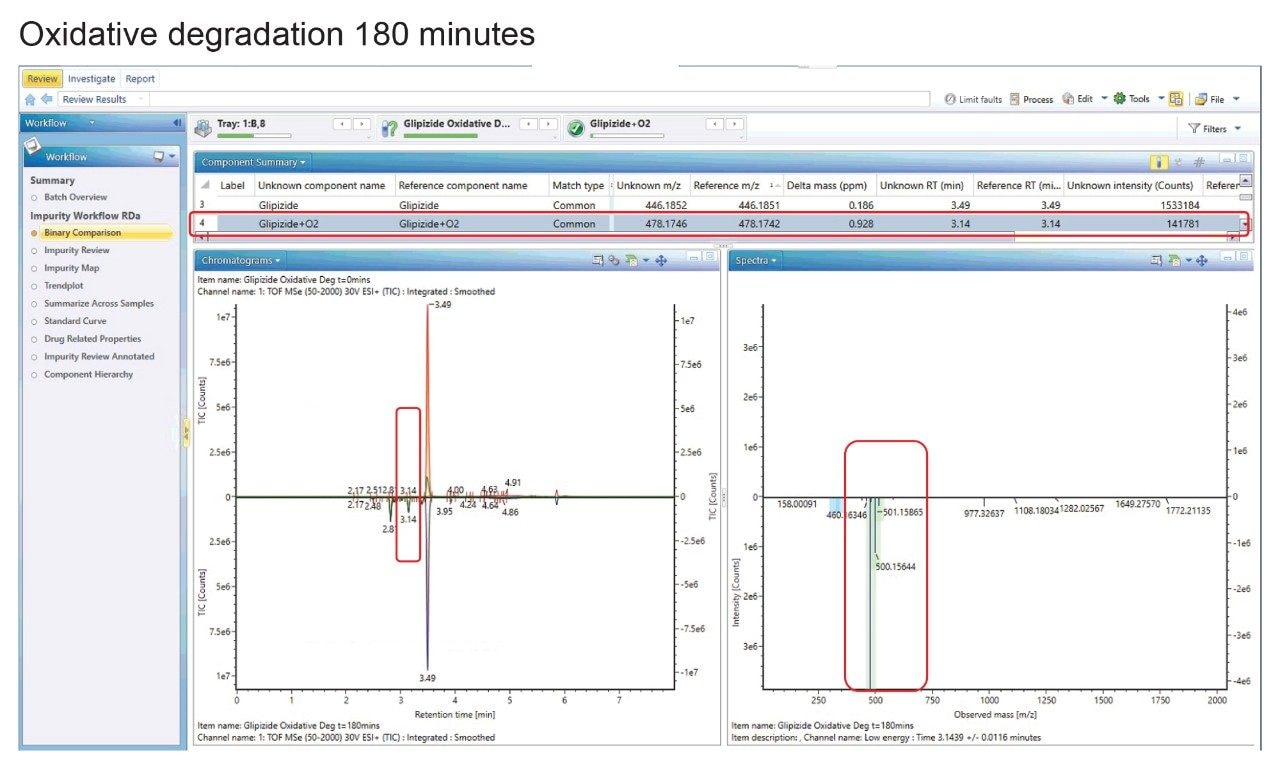
Binary Compare
For an initial appraisal of the unknown peak, again the Binary Compare function was used. In Figure 7 we have t=0 minutes for oxidative degradation as the reference and as the unknown, the t=180 minutes oxidative degradation (Figure 7). The peak highlighted in the ’targeted’ phase of the work is present only in the degraded sample at retention time 3.14 minutes. The peak at m/z 478.1746 was tentatively assigned as glipizide + O2
Drug-related impurities
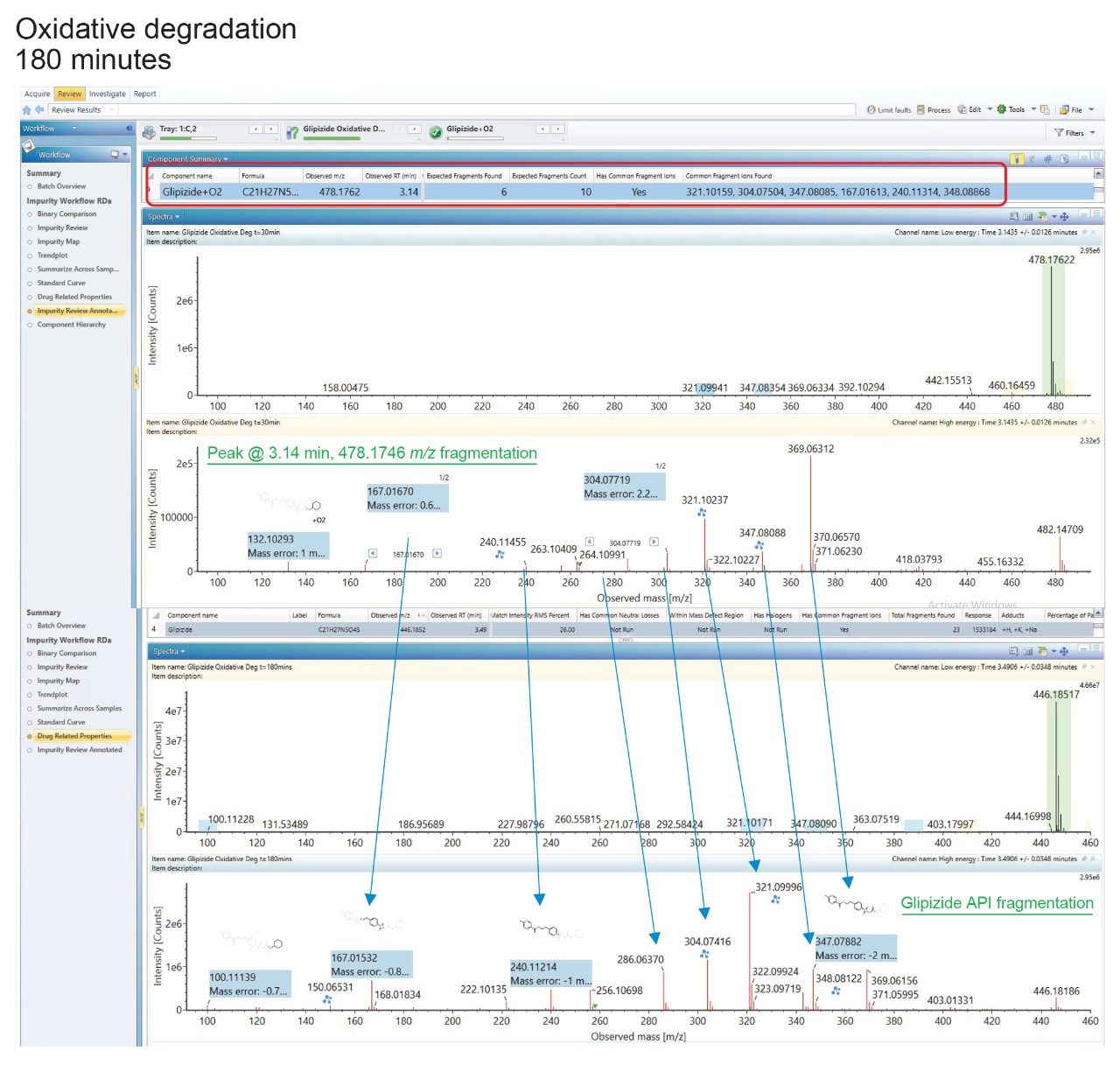
Further investigation of this suspected degradant was carried out to assess whether it was structurally related to the API. To this end, the product ions of the parent drug were added as detection results to the library entry for the API, then imported into the processing method.
The acquired data were screened against a method containing the parent compound and potential transformations based on the stress conditions, thereby identifying, and visualizing any related structures (Figure 8).
Six fragment ions, that had been automatically assigned by UNIFI from the structure of the API, were also detected in the high energy data of the suspected degradant and visualized. The charge-retaining portion is highlighted.
The presence of six fragment ions common to both the API and the suspected degradant strongly indicates the peak is structurally related to glipizide.
Conclusion
The ACQUITY RDa Detector coupled to the ACQUITY UPLC I-Class PLUS System and PDA Detector demonstrated the ability to provide targeted and non-targeted solutions to forced degradation studies.
The forced degradation of glipizide was successfully characterized using the waters_connect Software platform with UNIFI application. The workflows within UNIFI were able to automatically identify glipizide and corresponding degradation products with a high degree of confidence. The simultaneous acquisition of high energy data, generating fragment ions, provided additional structural information to aid in pathway profiling studies. All compounds and fragment ions presented exhibited excellent mass accuracy of less than 5ppm. This level of compound characterization normally requires expert users in HRMS to operate the instrument and interpret the results generated. Identification of unknowns would normally require outsourcing which can be expensive and time consuming to assess potential toxicity and/or negative impact on drug efficacy.
The waters_connect Software platform incorporating dedicated end-to-end workflows, combined with the ACQUITY RDa Detector’s automatic set-up, provides access to accurate mass measurement data in a routine workflow.
References
- Bansal, G, Singh, M, Jindal, KC, Singh, S. LC and LC-MS Study on Establishment of Degradation Pathway of Glipizide under Forced Decomposition Conditions. Journal of Chromatographic Science. 46:510–517; 2008.
- ICH guidelines, Q1A (R2): Stability Testing of New Drug Substances and Products (revision 2), International Conference on Harmonization. Available from: 〈http://www.fda.gov/downloads/RegulatoryInformation/Guidances/ucm128204.pdf〉, 2003.
- FDA Guidance for Industry, INDs for PhaseII and III Studies — Chemistry, Manufacturing, and Controls Information, Food and Drug Administration. Available from: 〈http://www.fda.gov/downloads/ Drugs/Guidance Compliance Regulatory Information/Guidances/ucm070567.pdf〉, 2003.
- Blessy, M, Ruchi D. Patel, Prajesh N Prajapati, Y.K. Agrawal. Development of Forced Degradation and Stability Indicating Studies of Drugs — A Review. Journal of Pharmaceutical Analysis 2014;4(3):159–165.
720007510, February 2022