Optimizing IP-RP CRISPR sgRNA Purification Passes With High Efficiency Oligonucleotide Certified BEH 300 Å C18 5 µm Preparative Sorbent
Abstract
CRISPR technology, which utilizes single-guide RNA (sgRNA) to target specific DNA sequences for gene editing, insertion, and epigenetic modifications, has emerged as a powerful tool in genetic engineering. Ensuring the high purity of sgRNA is critical to minimizing off-target effects and maximizing therapeutic precision. This study investigates the purification of a 100mer sgRNA using ion-pairing reverse-phase liquid chromatography (IPRP-LC) across various columns, with a particular focus on the performance capabilities of a newly commercialized oligonucleotide batch tested 5 µm BEH 300 Å C18 sorbent. Results demonstrate that the 5 µm preparative column significantly improves impurity separation and retention efficiency. The enhanced binding properties and loading capacity of the 5 µm Oligonucleotide BEH 300 Å C18 Column underscore its utility in high-resolution purification applications for CRISPR-related sgRNAs.
Benefits
- Batch tested and selected XBridge™ Oligonucelotide BEH C18 5 µm 300 Å sorbent particles for predictable, low secondary interaction adsorptive interactions
- Widepore organosilica sorbent columns with optimized diffusion kinetics for CRISPR sgRNA, chemical rugged for many repeat runs, and mechanically stability confered by specialized optimal bed density packing procedures
- Increases in accessible surface area per column length as a result of reduced particle size and pore size optimization
Introduction
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is a groundbreaking technology that has revolutionized the field of genetic engineering. It is primarily used for precise gene editing, gene insertion, and even epigenetic genome modifications.1 This cellular system relies on the use of a single-guide RNA (sgRNA) to direct a CRISPR-associated (Cas) protein, such as Cas9, to a specific DNA sequence. Once bound, the Cas protein induces a double-strand break, enabling either gene disruption or insertion. Beyond editing, CRISPR technology is now being utilized for epigenetic modifications to regulate gene expression without altering the DNA sequence itself, opening new avenues in therapeutic applications, disease modeling, and functional genomics.2
The application of CRISPR for gene editing and therapeutic interventions demands a high level of precision. The specificity of the sgRNA-Cas complex in targeting the intended sequence is crucial, as off-target effects can lead to unintended genetic modifications, which may compromise the safety and efficacy of the approach. Thus, the purity of sgRNA used in CRISPR experiments is of paramount importance. Impurities present in the sgRNA preparation can affect the binding efficiency and accuracy of the sgRNA-Cas complex, increasing the likelihood of off-target DNA cleavage events. Therefore, optimizing the purification process to remove impurities and ensure the highest possible sgRNA purity is vital for minimizing these risks.
The purification of sgRNA typically employs chromatographic techniques that exploit the physicochemical properties of nucleic acids.3 On a laboratory scale, ion-pairing reverse-phase (IPRP) liquid chromatography is a commonly used method, particularly effective for separating oligonucleotides like sgRNA based on their hydrophobic interactions with the stationary phase. IPRP is often chosen due to its ability to discriminate between full-length products and various truncated or chemically modified versions of sgRNA, which may arise during synthesis. sgRNA sequences are frequently prepared by solid-phase phosphoramidite chemical synthesis so that important synthetic modifications such as 2'-O-methylation or phosphorothioate linkages can be readily incorporated for enhanced stability and functionality. The crude sgRNA mixture, which contains both the desired full-length product and various synthesis by-products or impurities, is subjected to IPRP chromatography. Using specific buffer systems and optimized gradients, impurities can be separated based on retention time differences, allowing for the isolation of high-purity sgRNA fractions.
Traditionally, large particle size columns containing 8 to 20 µm diameter sorbents have been applied. However, to enhance separation efficiency and loading capacity of sgRNA, column characteristics such as pore size and particle size should be carefully considered. In this work, we therefore investigate the use of a new oligonucleotide batch tested and selected widepore BEH C18 sorbent. Herein, BEH C18 Columns with smaller particle sizes (e.g., 5 µm) and optimized pore dimensions (e.g., 300 Å) are applied to achieve new single pass, purity levels that are required for high-precision CRISPR applications.
Experimental
Sample Preparation
A 100mer sgRNA with sequence 5'-3 PTO 2'OMe RNA-25 natural RNA-12 2'OMe RNA-28 natural RNA-29 2'OMe RNA - 3 PTO 2'OMe RNA-3’ was used to demonstrate the differences in separation between three different purification columns, utilizing ion-pairing reverse phase purification. An analytical purity run on the crude starting material is provided below (Figure 1).
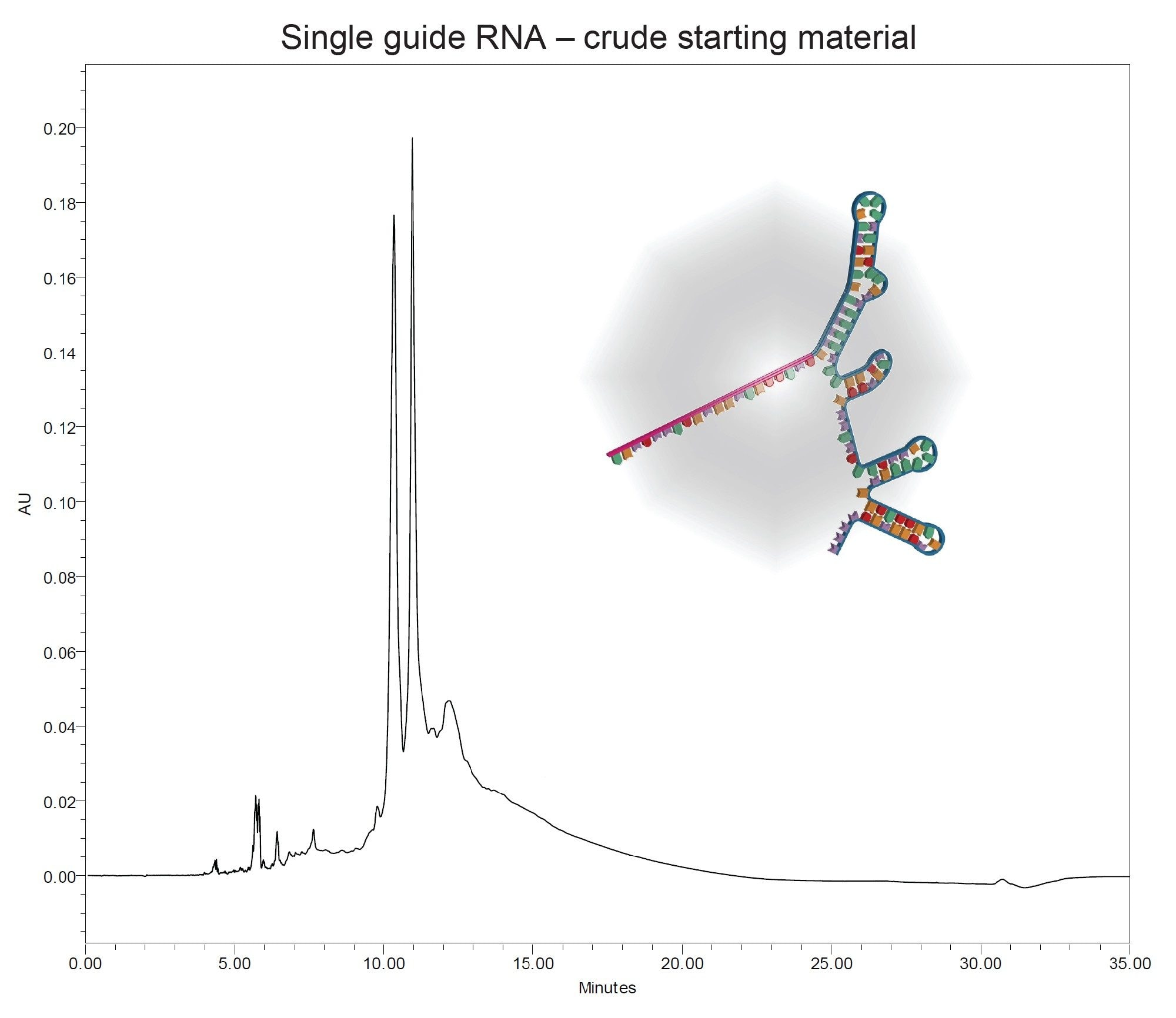
Previous work with sgRNA demonstrated better intensity and resolution using a combination of TEA and HFIP for analysis of sgRNA while Hexylammunium acetate (HAA) buffers were more effective for preparative scale. Due to absence of a direct scale up method, a screening gradient followed by a tailored gradient was applied to each column. The screening gradient determined the elution time for the purification column using HAA buffer. Based on this information, the tailored gradient was designed around the retention time of the full length product (FLP) identified during the screening gradient.
Purification IP-RPLC
|
LC system: |
Waters™ LC Prep 150 System, comprised of a 2545 Quaternary Solvent Manager, 2489 UV/Visible (UV/Vis) Detector™, and WFC III. Fraction collection was triggered by peak detection. |
|
Detection: |
Peak Detection – 0.1 AU |
|
Wavelength: |
275 nm |
|
Data acquisition: |
MassLynx™ |
|
Column: |
XBridge Peptide BEH C18 OBD Prep Column 300 Å, 10 µm, 19 mm x 250 mm (p/n: 186003673) XBridge Oligonucleotide BEH C18 OBD Prep Column 300 Å, 5 µm, 19 mm x 150 mm (p/n: 186011175) 25 x 150 mm 8 µm PS-DVB 300 Å Preparative Column (Vendor A) |
|
Column temperature: |
Room Temperature |
|
Sample temperature: |
Room Temperature |
|
Vials: |
50 mL Falcon |
|
Injection volume: |
Variable – dependent on amount of material to be injected |
|
Flow rate: |
20 mL/min |
|
Mobile phase A: |
100 mM Hexylammonium acetate (HAA), pH 7.0 |
|
Mobile phase B: |
50 mM HAA, 50% ACN, pH 7.0 |
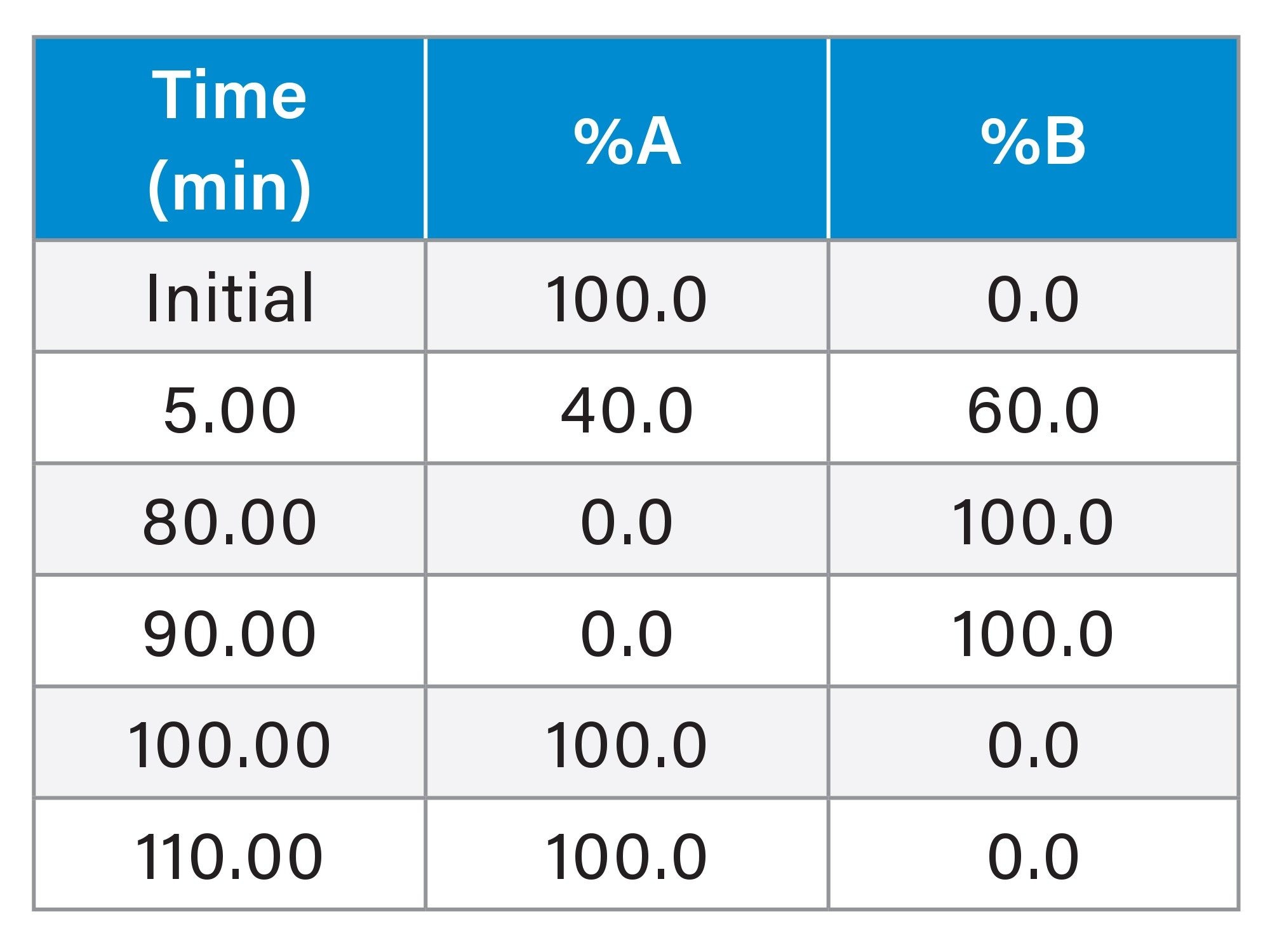
Gradient Table 1
Analytical IP-RPLC-UV/MS
|
LC system: |
Waters ACQUITY™ UPLC H-Class Plus System, comprised of BioQuaternary Solvent Manager, Bio Sample Manager FTN-H, and ACQUITY™ UPLC and ACQUITY Premier Tunable UV Detectors |
|
Wavelength: |
260 nm |
|
Data Acquisition: |
Empower™ |
|
Column: |
ACQUITY Premier Oligo BEH C18 300 Å, 1.7 µm 2.1 x 100 mm Column (p/n: 186010540) |
|
Column temperature: |
80 °C |
|
Injection volume: |
50 µL |
|
Flow rate: |
0.5 mL/min |
|
Mobile phase A: |
8 mM TEA, 80 mM HFIP in 100% water |
|
Mobile phase B: |
4 mM TEA, 40 mM HFIP in 50:50 water/MeOH |
|
TUV: |
260 nm |
|
Gradient: |
See Figure Caption |
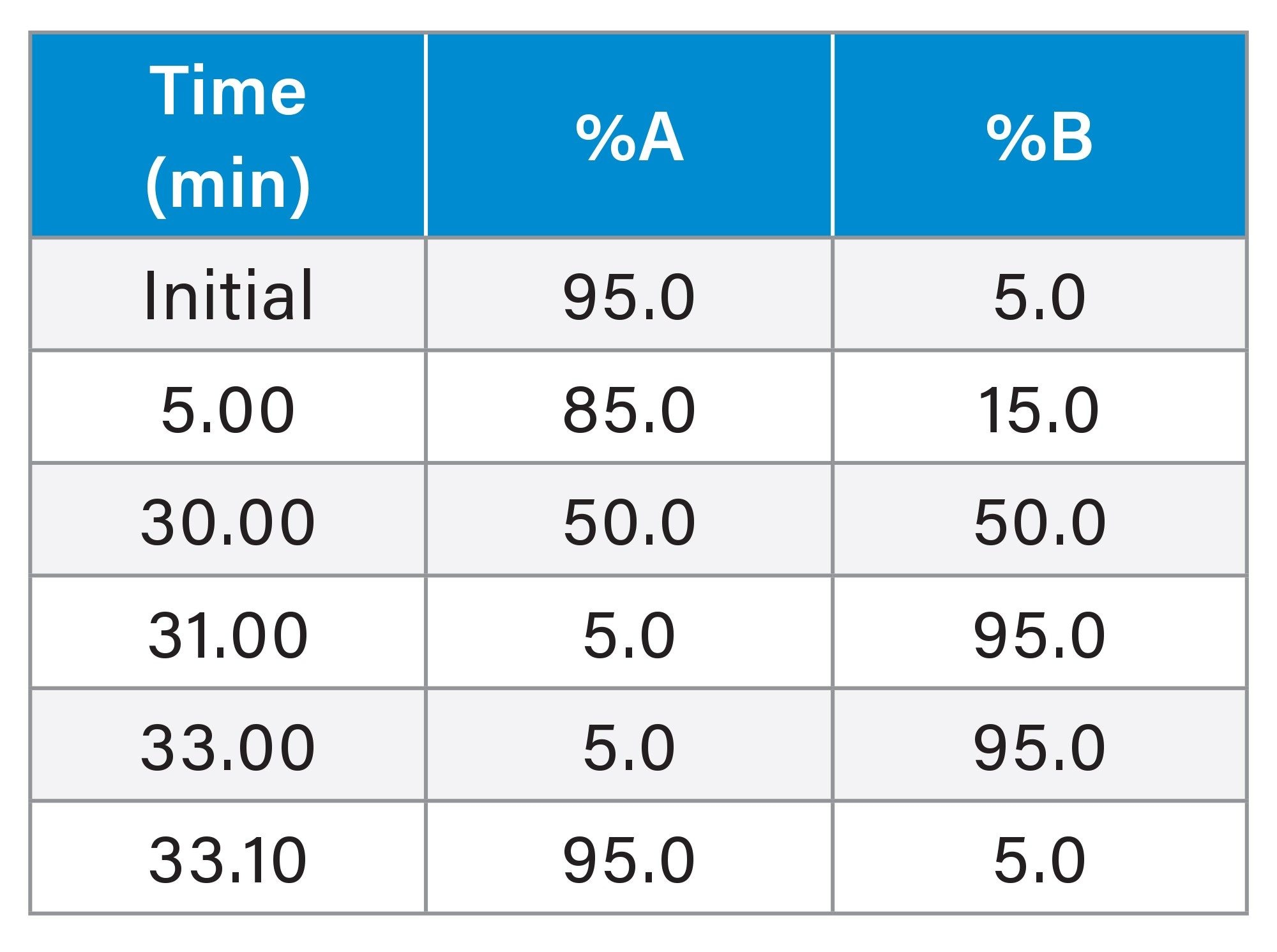
Gradient Table 2
Results and Discussion
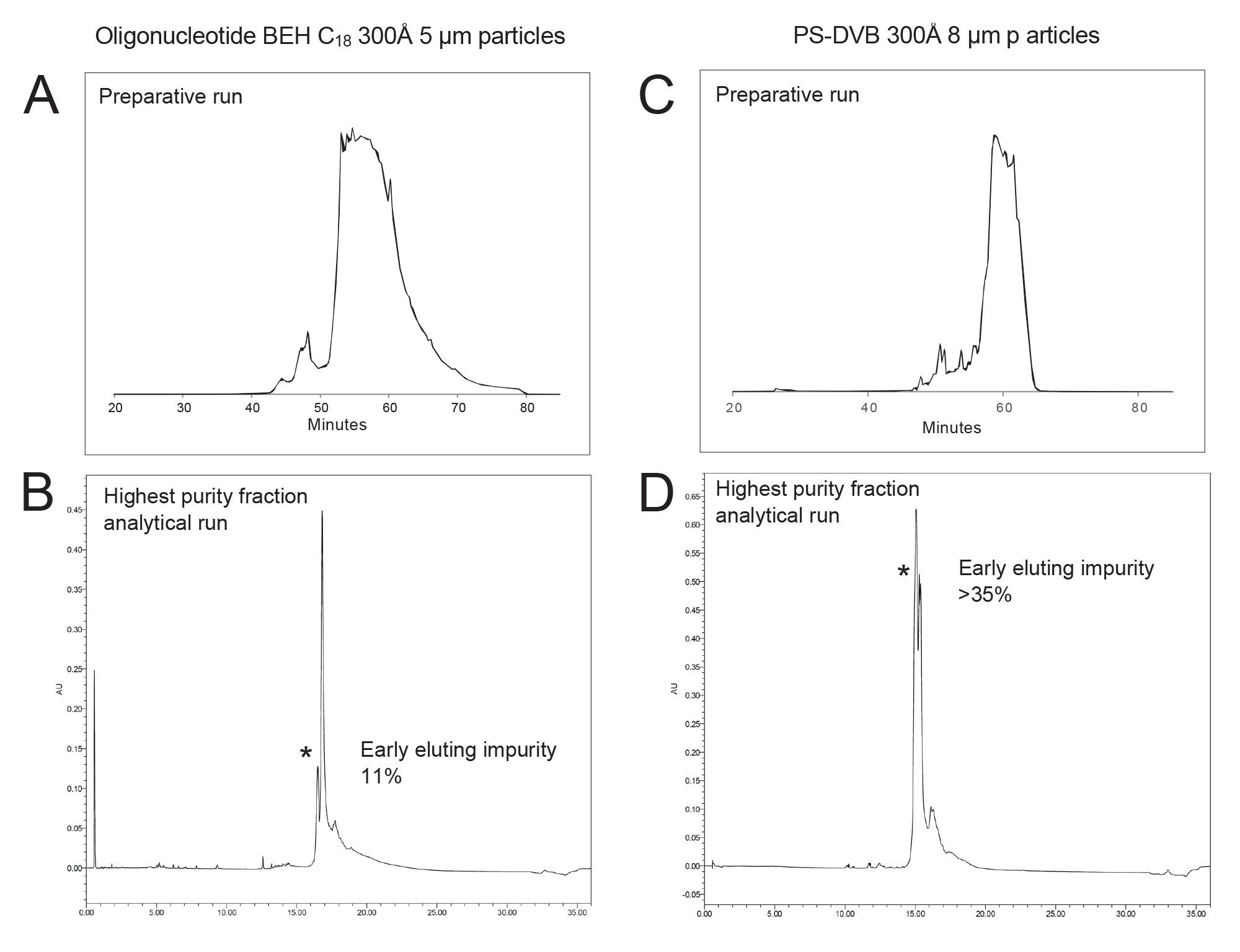
The purification of a 100mer sgRNA using various columns demonstrated significant performance differences. An sgRNA was purified through ion-pairing reverse-phase (IPRP) liquid chromatography using tailored gradients across multiple columns, including a 10 µm BEH 300 C18, 8 µm PS-DVB 300 Å, and a newly commercialized, batch tested and selected XBridge Oligonucleotide 5 µm BEH 300 C18 sorbent. Example purification runs obtained with both columns are shown in Figures 2A and 2C. The highest purity fractions collected from each was also analyzed and the corresponding analytical UV chromatograms are provided in Figures 2B and 2D, respectively.
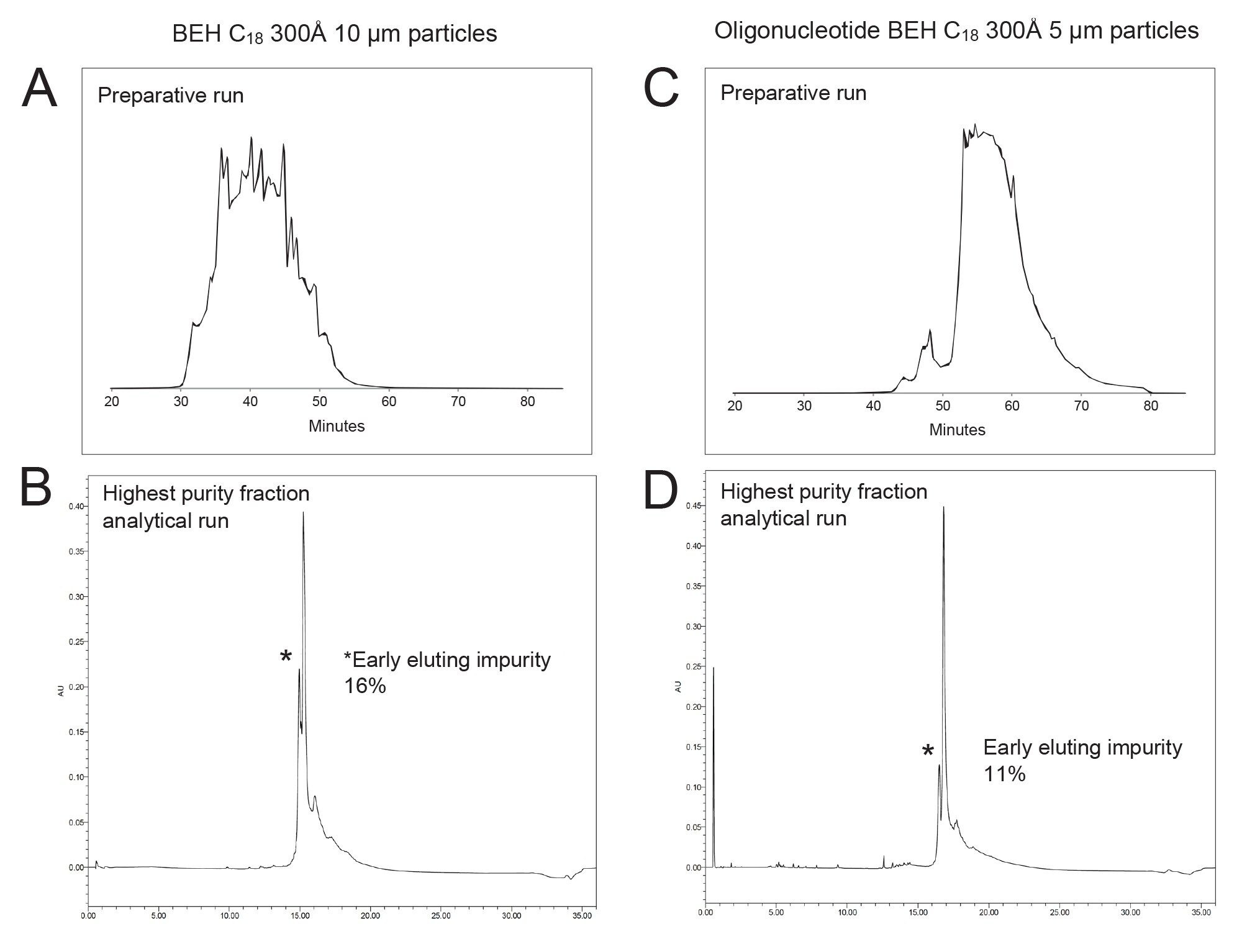
Assessing an XBridge Oligonucleotide BEH C18 OBD 300 Å 5 µm Column
The 5 µm Oligonucleotide BEH 300 Å C18 Column showed a marked improvement over its 10 µm counterpart in terms of purity and the separation of impurities. Initially, the sgRNA purification on the 10 µm column resulted in a purity level of ~35% (Best Fraction), with significant front impurity peaks (~16%) still present. Subsequent purification attempts failed to yield substantial improvements in separating these impurities, highlighting the limitations of the 10 µm sorbent in handling closely eluting species.
In contrast, the XBridge Premier Oligonucleotide BEH C18 300 Å 5 µm Column delivered superior performance across all purification rounds. The increased surface area provided by the smaller particle size of the 5 µm sorbent significantly enhanced the binding efficacy, resulting in improved retention and better separation of impurities. This column achieved purity levels of approximately 41–45% in repeated purifications, with a consistent reduction of front impurity peaks to around 11%, demonstrating its efficiency in removing early-eluting contaminants. The enhanced binding and retention characteristics of the 5 µm sorbent column also can be translated into a higher loading capacity. The ability of the sorbent to retain and effectively separate higher concentrations of the sample contributed to more efficient purification cycles. This is especially beneficial for handling complex biomolecules like sgRNAs, where higher loading often necessitates greater separation capabilities due to the increased potential for co-eluting impurities.
Comparison to an 8 µm PS-DVB 300 Å Polymeric Sorbent Preparative Column
The performance of the XBridge Oligonucleotide 5 µm BEH 300 Å C18 Column was also compared to that of an 8 µm PS-DVB 300 Å polymeric sorbent preparative column. We conducted the same series of separations using identical conditions and samples. The preparative run with the DVB column is provided in Figure 3. Fractions collected across the peak were analyzed as before. The highest purity fraction obtained on this column was found to contain early eluting impurities between 34 and 37%, suggesting there to be negligible purification. Curiously, the column also exhibited pressure fluctuations during the purification.
Future Directions
Despite the clear advantages in removing front impurities, the purification process encountered challenges with late-eluting contaminants. These impurities are likely linked to incomplete deprotection of the natural RNA bases, an issue identified as a bottleneck in achieving full-length product purity. The purification efficiency for these impurities remained limited, even with the 5 µm BEH Column. Therefore, further optimization in sgRNA synthesis and deprotection processes is necessary to enhance the overall efficacy of the purification protocol. Moreover, in future work, we will apply concave gradients to improve the selectivity and overall run efficiency for separating out larger impurity components.
Conclusion
These results demonstrate that the XBridge Oligonucleotide BEH C18 300 Å 5 µm OBD Column provides significant advantages over larger particle size options, particularly in enhancing impurity separation and increasing loading capacity. These benefits underscore its utility in the high-resolution purification of sgRNAs and similar oligonucleotide products, emphasizing its role as a preferred choice in high-performance liquid chromatography (HPLC) methods. Future investigations will focus on addressing the late-eluting impurities through synthesis optimization and exploring alternative gradients and purification schemes that combine weak and strong ion pairing passes as well as anion exchange coupled with IP-RP purification.
References
- Villiger, L., Joung, J., Koblan, L. et al. CRISPR technologies for genome, epigenome and transcriptome editing. Nat Rev Mol Cell Biol 25, 464–487 (2024). https://doi.org/10.1038/s41580-023-00697-6.
- Nakamura, M., Gao, Y., Dominguez, A.A. et al. CRISPR technologies for precise epigenome editing. Nat Cell Biol 23, 11–22 (2021). https://doi.org/10.1038/s41556-020-00620-7.
- Minkner R, Boonyakida J, Park EY, Wätzig H. Oligonucleotide separation techniques for purification and analysis: What can we learn for today's tasks?Electrophoresis. 2022; 43: 2402–2427. https://doi.org/10.1002/elps.202200079.
720008641, December 2024